Abstract
Differential analysis of gene and transcript expression using high-throughput RNA sequencing (RNA-seq) is complicated by several sources of measurement variability and poses numerous statistical challenges. We present Cuffdiff 2, an algorithm that estimates expression at transcript-level resolution and controls for variability evident across replicate libraries. Cuffdiff 2 robustly identifies differentially expressed transcripts and genes and reveals differential splicing and promoter-preference changes. We demonstrate the accuracy of our approach through differential analysis of lung fibroblasts in response to loss of the developmental transcription factor HOXA1, which we show is required for lung fibroblast and HeLa cell cycle progression. Loss of HOXA1 results in significant expression level changes in thousands of individual transcripts, along with isoform switching events in key regulators of the cell cycle. Cuffdiff 2 performs robust differential analysis in RNA-seq experiments at transcript resolution, revealing a layer of regulation not readily observable with other high-throughput technologies.
RNA-seq is a high-throughput sequencing assay that can be used for both discovery and quantification of transcripts in a single experiment1–4. Recent studies have shown RNA-seq to be more accurate over a larger dynamic range of gene expression than expression microarrays5,6. Relating genetic variation and epigenetic state to transcriptional and post-transcriptional regulation is a major goal in many large-scale genomic analyses. RNA-seq has become a vital component in these efforts, several of which have implicated alternative splicing and isoform selection as determinants of cell type and specificity7–12. Moreover, many genes have a large genomic ‘footprint’, with numerous splice variants, promoters and protein products. Determining how isoform selection and diversity is regulated requires measuring changes in the expression of individual transcripts. In this work we describe an algorithm to robustly track the dynamics of transcript expression. (We use transcript and isoform interchangeably, and refer to a single RNA species that may be the product of an alternatively spliced gene. Gene refers to a set of one or more transcripts that share some amount of sequence in common.)
Current RNA-seq differential analysis methods focus on tackling one of two major challenges. The first is accurately deriving gene and isoform expression values from raw sequencing reads, which requires statistical computations at isoform-level resolution3,13–16. The second is accounting for variability in measurements across biological replicates of an experiment17–22. To our knowledge, no algorithm has rigorously addressed both problems simultaneously for genes and transcripts. Although in some cases methods for measurement of expression at isoform-level resolution partly address the differential analysis problem3,14,23, they ignore the issue of variability across biological replicates, leading to over-prediction of differentially abundant transcripts and high false-positive rates. A recent method modeled variability in transcript-level but not gene-level expression24. Methods to control for variability in gene expression across replicates have been focused mainly on controlling for variability in the raw read data, but they miss key aspects of accurately transforming reads into gene expression values. Alternative splicing and repetitive regions introduce uncertainty into gene expression measurements, and failing to control for this uncertainty can introduce errors during differential analysis. A recent method for assessing differential splicing was focused on biological variability in inclusion rates for individual exons, but the approach did not extend to complete transcripts25. Thus, methods for differential analysis of RNA-seq have yet to accurately control for key sources of variability at gene- and transcript-level resolution simultaneously, and therefore do not realize the full potential of the assay to capture transcriptome dynamics.
Here we introduce Cuffdiff 2, which addresses both problems simultaneously by modeling variability in the number of fragments generated by each transcript across replicates. (We use the term fragment to refer to an RNA-Seq library fragment, which may be sequenced at one or both ends.) Cuffdiff 2 generates more accurate transcript-resolution estimates of changes in gene expression, compared with existing approaches, and is accurate over a wide range of RNA-seq designs, including those done on benchtop sequencers such as the Illumina MiSeq. We use Cuffdiff 2 to assess the response to knockdown of HOXA1, a member of a highly conserved family of transcription factors that establish body plan organization during development26. We show that HOXA1 is required for the survival of adult fibroblasts and HeLa cells. Cuffdiff 2 identified genes that displayed altered inclusion of key features, such as DNA binding regions, in their protein products.
Results
Raw fragment counts inaccurately estimate changes in expression
Early methods for quantifying gene expression from RNA-seq data work by counting the sequencing library fragments that map to the exons of each gene and dividing the count for each gene by a scaling factor based on the length of the exons. Expression levels estimated using such approaches are less accurate than later methods27, which calculate a gene's expression level by adding the expression values of its alternative isoforms3,16. We refer to the former as ‘raw count’ methods and the latter as ‘isoform deconvolution’ methods. Current tools for differential gene expression analysis use the raw count method, equating the change in a gene's expression levels with the change in the number of fragments originating from it between conditions17,20,21,28.
Because the raw count method is not always accurate when calculating gene expression in a single library, we hypothesized that it would be inaccurate when comparing libraries. Simple examples of hypothetical, alternatively spliced genes showed that the change in expression could be drastically different from the change in raw read count (Fig. 1). We compared expression levels from two popular raw count schemes to changes in gene expression in simulation experiments. When all of a gene's isoforms are up- or downregulated between two conditions, raw count methods recover true change in gene expression. However, when some isoforms are upregulated and others downregulated, raw count methods are inaccurate (Supplementary Fig. 1). In contrast, gene expression levels calculated by isoform deconvolution correlated well with true gene expression even when relative abundance of the isoforms changed between conditions. Thus, identifying accurate, statistically significant expression changes at the resolution level of genes requires transcript-level calculations.
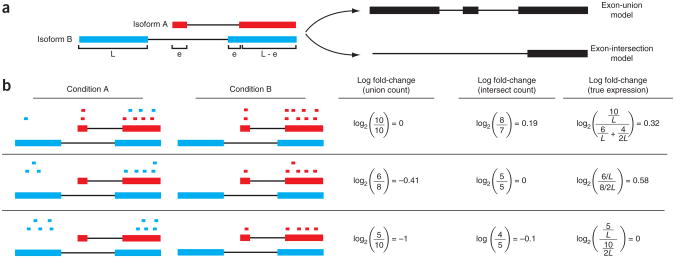
Figure 1.
Changes in fragment count for a gene does not necessarily equal a change in expression. (a) Simple read-counting schemes sum the fragments incident on a gene's exons. The exon-union model counts reads falling on any of a gene's exons, whereas the exon-intersection model counts only reads on constitutive exons. (b) Both of the exon-union and exon-intersection counting schemes may incorrectly estimate a change in expression in genes with multiple isoforms. The true expression is estimated by the sum of the length-normalized isoform read counts. The discrepancy between a change in the union or intersection count and a change in gene expression is driven by a change in the abundance of the isoforms with respect to one another. In the top row, the gene generates the same number of reads in conditions A and B, but in condition B, all of the reads come from the shorter of the two isoforms, and thus the true expression for the gene is higher in condition B. The intersection count scheme underestimates the true change in gene expression, and the union scheme fails to detect the change entirely. In the middle row, the intersection count fails to detect a change driven by a shift in the dominant isoform for the gene. The union scheme detects a shift in the wrong direction. In the bottom row, the gene's expression is constant, but the isoforms undergo a complete switch between conditions A and B. Both simplified counting schemes register a change in count that does not reflect a change in gene expression.
Cuffdiff 2
Cuffdiff 2 assumes that the expression of a transcript in each condition can be measured by counting the number of fragments generated by it. Thus, a change in the expression level of a transcript is measured by comparing its fragment count in each condition. If the chance of seeing a change in this count is small enough under an appropriate statistical model of the inherent variability in this count (say with odds of 1 in 100), the transcript is deemed significantly differentially expressed. Choosing a model that adequately controls for variability in sequencing depth, biological noise and splicing structure has been the subject of debate19. Under one of the simplest models, the Poisson model, the variability is estimated by calculating the mean count across replicates, which allows one to calculate a P-value for any observed changes in a transcript's fragment count.
The Poisson model is computationally simple, but it fails to account for two key issues that arise in differential analysis—count uncertainty and count overdispersion. Count uncertainty refers to the observation that in RNA-seq experiments it is common for up to 50% of reads to map ambiguously to different transcripts29. This happens because in higher eukaryotes alternative isoforms of most genes share large amounts of sequence, and many genes have paralogs with high sequence similarity. As a result, the fragment counts for individual transcripts cannot be calculated exactly and must be estimated. Count overdispersion refers to the fact that experiments that produce count data are often more variable across replicates than what is expected according to a Poisson distribution17,20.
Our method (Fig. 2) addresses both of these issues by modeling how variability in measurements of a transcript's fragment count depends on both its expression and its splicing structure. Previous studies observed that overdispersion in RNA-seq experiments increases with expression and proposed the negative binomial distribution as a means of controlling for it17,22. In contrast, ambiguity in mapping fragments to transcripts manifests itself in measurement uncertainty and error, and depends largely on splicing structure. Cuffdiff 2 determines the degree of overdispersion in this mixture by globally fitting the observed variance in fragment counts as a function of the mean across replicates (Supplementary Fig. 2). The algorithm then estimates the number of fragments that originated from each transcript, as previously described3,30. Next, it combines the uncertainty in each transcript's fragment count with the overdispersion predicted to exist for that count by the global model of cross-replicate variability. Cuffdiff 2 estimates uncertainty by calculating the confidence that each fragment is correctly assigned to the transcript that generated it; transcripts with more shared exons and few uniquely assigned fragments will have greater uncertainty. The algorithm captures uncertainty in a transcript's fragment count as a beta distribution and the overdispersion in this count with a negative binomial, and mixes the distributions together. The resulting mixture is a beta negative binomial distribution that reflects both sources of variability in an isoform's measured expression level.
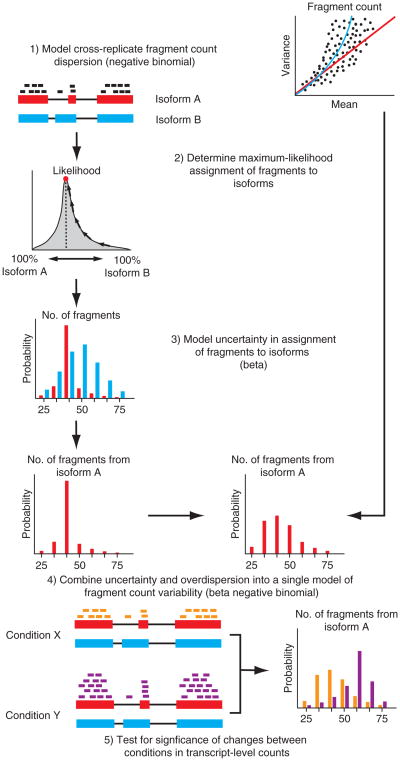
Figure 2.
An overview of the Cuffdiff 2 approach to isoform-level differential analysis of RNA-seq data. (1) The variability in fragment count for each gene across replicates is modeled. (2) The fragment count for each isoform is estimated in each replicate, along with (3) a measure of uncertainty in this estimate arising from ambiguously mapped reads, which are extremely prevalent in alternatively spliced transcriptomes. (4) The algorithm combines estimates of uncertainty and cross-replicate variability under a beta negative binomial model of fragment count variability to estimate count variances for each transcript in each library. (5) These variance estimates are used during statistical testing to report significantly differentially expressed genes and transcripts.
Cuffdiff 2 estimates expression at gene- and transcript-level resolution, the variance in the expression levels and the covariances between isoforms of the same gene from replicate experiments. This allows it to accurately estimate gene expression and perform differential analysis at gene-level resolution without encountering the limitations inherent in the raw count methods discussed above. The software reports to the user the change in expression for each gene and transcript, along with statistical significance scores for these changes.
Response to loss of HOXA1 at gene- and transcript-level resolution
To demonstrate the effectiveness of transcript-resolution RNA-seq analysis, we selected a biological problem arising from an ongoing study of the role of HOX gene function in adult cells. Genes in the HOXA cluster, which are critical for proper body patterning during development, have spatial expression patterns in adult cells that identify their anatomic origin31. Whether this expression pattern is functionally relevant in adult cell types has been so far unanswered.
We performed RNA interference (RNAi)-mediated knockdown of HOXA1 in human primary lung fibroblasts, where HOXA1 was depleted using a pool of four short interfering RNAs (siRNAs) targeting HOXA1 designed to minimize off-target effects. We controlled for a nonspecific RNAi response by comparing HOXA1-depleted fibroblasts against cells treated with a pool of scrambled siRNAs that do not target a specific gene. We isolated total RNA in biological triplicate 48 h after transfection. Sequencing of the poly-A–selected fraction on an Illumina HiSeq 2000 yielded >231 million 100-bp paired-end RNA-seq reads. The same RNA was labeled and hybridized to Agilent SurePrint G3 Gene Expression arrays (Online Methods).
Cuffdiff 2–derived changes in gene expression in response to HOXA1 knockdown strongly agreed with values from microarrays (Spearman correlation = 0.85), consistent with previous comparisons2,5 (Fig. 3a and Supplementary Fig. 3). Changes in multi-isoform gene expression calculated by Cuffdiff 2 improved concordance with the array measurements by 15% compared with the change in raw count (Fig. 3b and Supplementary Figs. 4 and 5). The discrepancy between raw count and Cuffdiff 2 measurements of gene expression tended to be higher for genes where alternative isoforms shift in expression relative to one another, a phenomenon we term ‘isoform switching’. (Supplementary Figs. 6 and 7).
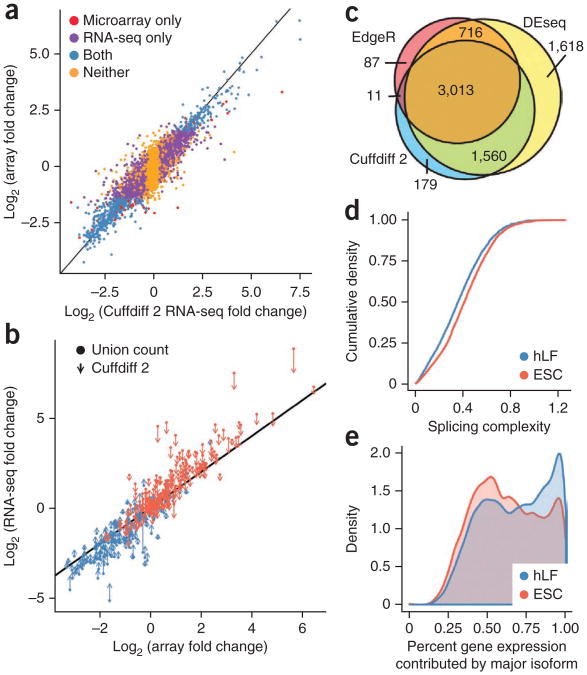
Figure 3.
Comparison of Cuffdiff 2 with other expression platforms. (a) Fold changes in multi-isoform gene expression measured by RNA-seq and microarrays before and after HOXA1 knockdown are highly concordant (Spearman correlation = 0.86). (b) Computing gene expression by isoform deconvolution instead of by fold change in gene-level fragment counts improves agreement with microarrays. Genes shown are those where Cuffdiff 2 and intersection-count fold changes are most discrepant (1% tails). (c) Methods for performing differential gene expression analysis with RNA-seq based on the exon-union counting method returned nearly all of the genes returned by Cuffdiff 2. (d) Lung fibroblasts show lower splicing complexity than human ESCs. Complexity is measured by the Shannon-Entropy of the relative isoform abundances of multi-isoform genes (Methods), where zero complexity indicates a gene has only a single detectable isoform. A gene with equally abundant isoforms has maximal complexity. hLF, human lung fibroblast. (e) The contribution of the major isoform of each gene to total gene expression in lung fibroblasts and ESCs.
Cuffdiff 2 returned far more statistically significant differentially expressed genes than microarray analysis. Cuffdiff 2's differentially expressed genes contained 623 of the 745 (84%) reported by the arrays, along with an additional 4,138 genes (false-discovery rate (FDR) <1%). Moreover, Cuffdiff 2 was highly concordant with the popular count-based tools, with >94% of genes reported as differentially expressed also identified by the popular raw-count methods DESeq or edgeR (Fig. 3c).
Cuffdiff 2 detected expression for 16,278 of 69,202 (38%) transcripts in the annotated transcriptome (UCSC hg19 coding genes; http://genome.ucsc.edu/), and identified an average of 1.15 differentially expressed transcripts per differentially expressed gene in response to loss of HOXA1. Alternative isoform abundances relative to one another were maintained in most genes, with only 170 genes undergoing significant (FDR ≤ 1%) differential splicing, coding DNA sequence use or promoter preference. Splicing complexity has been reported to be higher in stem cells32 than in differentiated cell types, and through isoform ‘specialization’ to be reduced during lineage commitment. We compared human lung fibroblasts to human embryonic stem cells (ESCs). Cuffdiff 2 detected more differentially expressed transcripts (1.35 differentially expressed transcripts per differentially expressed gene) and higher overall splicing complexity than in lung fibroblasts. The relative abundance of major isoforms in ESCs contributed less to overall gene expression than major isoforms in fibroblasts, consistent with increased isoform specialization in fibroblasts (Fig. 3d,e).
Cuffdiff 2 is accurate over a wide range of experimental designs
To establish the accuracy of expression changes reported by Cuffdiff 2, we next analyzed published RNA-seq data with matched quantitative (q)PCR measurements. Analysis of the microarray quality control data confirmed that Cuffdiff 2, DESeq and edgeR all produce accurate measures of fold change. DESeq and edgeR report more genes overall as being differentially expressed than does Cuffdiff 2, most of which have small fold changes or very low sequencing depth, consistent with Cuffdiff 2's control for variance in expression owing to fragment count uncertainty. On another RNA-seq data set, Cuffdiff 2 returned changes in transcript-level expression concordant with both RSEM and ALEXA-Seq, which estimate isoform expression levels in individual RNA-seq samples, and confirmed by a matched exon-specific qPCR data set (Supplementary Figs. 7–10).
Depth of sequencing, read length and number of experimental replicates are major determinants of cost in RNA-seq experiments. To assess tradeoffs in accuracy versus cost under different experimental designs, we designed a test framework to simulate sequencing. We perturbed 1,000 randomly selected multi-isoform genes from the fibroblast data by modulating expression of individual transcripts and then simulated RNA-seq before and after perturbation. The simulations included scenarios that involved low, moderate and heavy levels of isoform switching, comparable to real contrasts (Online Methods and Supplementary Fig. 11). Cuffdiff 2 accurately recovered differentially expressed genes and transcripts across a range of read lengths, and replication and sequencing depths (Fig. 4a, Supplementary Tables 2,3 and Supplementary Figs. 12–76).
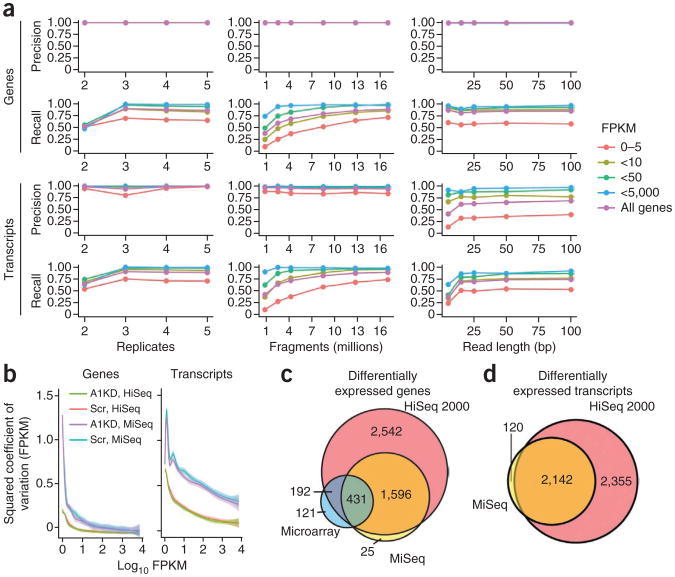
Figure 4.
Accuracy of Cuffdiff 2 over varied experimental designs. (a) Accuracy of the proposed model explored through simulated RNA-seq. Read-length series was generated with single-ended sequencing data. FPKM, fragments per kilobase per million fragments mapped. (b) Squared coefficient of variation versus expression for genes and transcripts as measured by the HiSeq 2000 and MiSeq. Each series is a fit of a generalized additive model to the individual expression, squared coefficient of variation pairs. (c) Significant gene lists returned by Cuffdiff 2 using the HiSeq 2000 and MiSeq compared against expression microarrays (FDR ≤ 1%). (d) Significant isoform lists returned by Cuffdiff 2 when using the HiSeq 2000 and MiSeq (FDR ≤ 1%).
An alternative method for performing transcript-level differential analysis would be to estimate the number of fragments originating from each transcript in each replicate library using Cufflinks or RSEM, and then provide these counts as input to a count-based tool such as DESeq or edgeR. Although these tools were not designed to be used in this way, and doing so violates some key assumptions of the underlying statistics assumed by both tools, their variance modeling procedures are flexible and robust.
In simulations designed to match our fibroblast experiment, Cuffdiff 2 outperformed alternative workflows that combine estimated transcript counts with DESeq or edgeR, consistent with a similar assessment of this approach24. (Supplementary Fig. 77). All methods detected >70% of differentially expressed transcripts. DESeq detected differentially expressed transcripts at similar sensitivity to Cuffdiff 2 but returned ∼125 false-positive transcripts. Although edgeR returned fewer false positives (∼75) than DESeq, it also detected fewer true positives than Cuffdiff 2. False positives were concentrated in genes with complex splicing and multiple highly similar isoforms. In contrast, Cuffdiff 2 returned ∼50 false positives and the false-positive rate was not noticeably worse for genes with many isoforms, indicating that Cuffdiff 2 performs robust transcript-level differential analysis of even very complex gene annotations.
Cuffdiff 2 had high precision (≥99%) at gene-level resolution across the dynamic range of expression, generating few false positives regardless of replication depth. DESeq and edgeR returned slightly more false-positive differentially expressed genes than Cuffdiff 2, except for scenarios with substantial isoform switching between conditions, where these tools had high FDRs. At transcript-level resolution, Cuffdiff 2's precision was 95–99% over nearly all designs. Adding additional replicates reduced false-positive transcripts returned by Cuffdiff 2, nearly eliminating them with five in each condition. In contrast, providing DESeq or edgeR with transcript-level fragment count generated ∼100 false positives even with five replicates. The detection rate (recall) of differentially expressed genes and transcripts was similar for three or more replicates, and lower for fewer replicates, due to a more conservative dispersion modeling mode used by Cuffdiff 2 for these designs (Online Methods).
The detection rate of differentially expressed genes and transcripts strongly depended on sequencing depth. However, recall of differentially expressed genes and transcripts in the top three quartiles of expression suffered surprisingly little even at 4 million fragments, constituting 25% of the starting sequencing depth. Importantly, the false-positive rate held constant across all depths. Recall suffered at lower depths for transcripts in the bottom quartile of expression, suggesting that deep sequencing is critical for differential analysis of genes present at low abundance, such as long noncoding RNAs33.
In contrast to the striking impact of sequencing depth, long or paired reads provided a modest improvement in accuracy for gene- and transcript-resolution analysis. Nevertheless, because these data were obtained with simulated reads that were perfectly mapped to the genome, we caution against designs with short (<75 bp) unpaired reads, as errors in upstream mapping and transcriptome assembly analyses are more common with such data.
For nearly all conditions tested, FDR was controlled at or below the target level. An independent assessment of FDR obtained by comparing ‘nonsense’ groups of replicates from real sequencing experiments confirmed that Cuffdiff 2 reports few false-positive differentially expressed transcripts and genes, and few genes undergoing isoform switching (Supplementary Table 1). Cuffdiff 2 more robustly captured gene- and transcript-level changes than count-based tools even for conditions with high cross-replicate variability or dramatic isoform switching (Supplementary Figs. 53–72).
Differential transcriptome analysis on a benchtop sequencer
Notably, the simulation results predicted that Cuffdiff 2 could recover transcriptome dynamics from the HOXA1 knockdown with far less data than generated by the HiSeq 2000. Recently available ‘bench-top’ sequencers such as the MiSeq (Illumina) and the Ion PGM Sequencer (Ion Torrent Systems) promise to broaden availability of RNA-seq for differential analysis, but whether these instruments yield enough data for accurate quantification is an open question.
Sequencing the HOXA1 and knockdown RNA-seq libraries from fibroblasts across three runs of a MiSeq produced 54 million paired reads (23% of the HiSeq data). Empirical estimates of cross-replicate variance in expression were substantially higher for genes and transcripts (Fig. 4b and Supplementary Figs. 78–81), but Cuffdiff 2 still recovered nearly 50% of the differentially expressed genes and transcripts from the HiSeq 2000 (Fig. 4c,d). Importantly, few genes and transcripts were reported as differentially expressed with the MiSeq data but not the HiSeq data, confirming that Cuffdiff 2 does not generate excess false positives at lower sequencing depth.
HOXA1 is required for viability in HeLa cells and lung fibroblasts
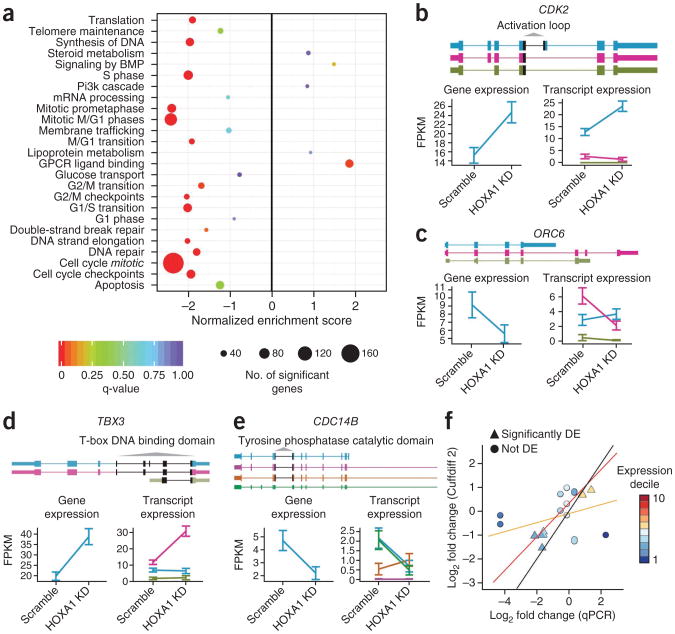
To further assess the biological insight gleaned from the transcript level response to loss of HOXA1, we performed Gene Set Enrichment Analysis (GSEA)34. The 15 gene sets most strongly downregulated in response to loss of HOXA1 contained hundreds of genes known to be specific to S phase, mitosis and the G2/M phase (Fig. 5a). Moreover, numerous genes in the p53 pathway, including p15 and p21, were upregulated. Cyclins A2, B1, B2, C, E2 and F were significantly reduced (FDR < 0.01%) in response to loss of HOXA1 (Supplementary Fig. 82). These data are consistent with inhibition of cell cycle progression at the G1-S transition, induction of apoptosis or a mixture of both.
Figure 5.
Changes in expression of cell cycle regulatory genes in response to HOXA1 knockdown. (a) GSEA analysis of the knockdown for selected REACTOME gene sets. (b) Cuffdiff 2 reports an increase in CDK2 expression, which is attributable to a single isoform that includes the full activation loop, a feature required for maximal CDK2 activity. KD, knockdown. (c) Cuffdiff 2 reports a decrease in ORC6 expression, which is attributable to a single isoform that includes the full suite of residues required for optimal DNA binding not present in the minor isoforms arising from the gene. (d) Cuffdiff 2 reports an increase in TBX3 attributable to a single isoform lacking an exon situated within the T-box DNA binding domain that is present in a highly similar minor isoform. (e) Cuffdiff 2 reports a decrease in CDC14B attributable to decreases in the two major isoforms. Error bars indicate 95% confidence intervals in expression. (f) Changes in isoform expression reported by Cuffdiff 2 compared against measurements made with isoform-specific qPCR. The black line indicates perfect correspondence between the two platforms. The orange line is a linear regression through all points, and the red line excludes the three major outliers, which target low abundance isoforms, two of which cannot be distinguished from primary transcript or genomic DNA. DE, differentially expressed.
We next investigated significant shifts in relative isoform usage identified by Cuffdiff 2. Cuffdiff 2 identified 170 genes that underwent significant shifts (FDR ≤ 1%) in transcript usage, including key regulators of cell cycle progression (ORC6, CDC14B) and gene expression (e.g., TBX3)35–40 (Supplementary Fig. 83). Close examination of specific isoform-switching events revealed that in some instances, the changes could induce differential production of key protein sequence features or domains. Only one of three isoforms of CDK2, a G1-S phase–specific regulatory kinase, is upregulated in response to loss of HOXA1. The upregulated transcript includes an exon that alters the length and amino acid sequence of the activation loop, which confers specificity for cyclin binding and substrate phosphorylation35 (Fig. 5b). Likewise, the DNA binding domains of the TBX3 transcription factor and the origin of recognition complex component ORC6 could be altered by means of isoform switching in response to HOXA1 reduction, possibly altering their roles in cell proliferation and survival36 (Fig. 5c–e). Isoform-specific qPCR confirmed the changes in isoform expression reported as significant for these genes (Fig. 5f and Supplementary Figs. 84 and 85).
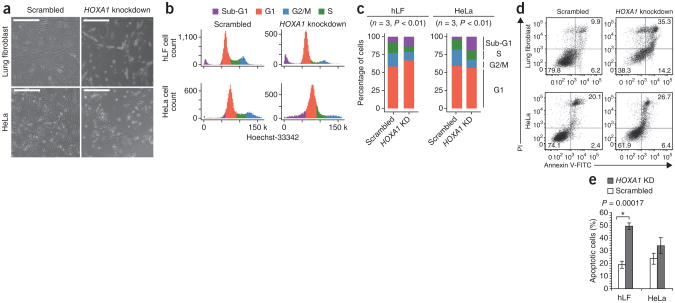
To test whether HOXA1 plays a role in cell cycle progression and viability in different human cell types, we also depleted HOXA1 in HeLa cells. Similar to what was observed in primary fibroblasts, cell numbers were drastically reduced in HeLa by 72 h after siRNA treatment relative to nontargeting controls (Fig. 6a). Bioinformatic analysis and transfection of multiple independent siRNAs excluded off-target knockdowns as the source of this phenotype (Supplementary Figs. 86 and 87). We next performed cell cycle analyses following loss of HOXA1 and observed a block at the G1 phase of the cycle in both cell types. This was also associated with an increase in the sub-G1 fraction (Fig. 6b,c), which was consistent with increased apoptosis as assessed by Annexin V staining (Fig. 6d,e). Together, these data offer functional evidence that loss of HOXA1 impairs cellular progression through the G1 phase and leads to apoptosis in these cell types, as suggested by Cuffdiff 2.
Figure 6.
Cell cycle analysis after HOXA1 knockdown. (a) Human lung fibroblasts transfected with scrambled siRNAs and a HOXA1 siRNA pool 48 h after transfection. HeLa cells transfected with scrambled siRNAs (left) and a HOXA1 siRNA pool 72 h after transfection (right). Scale bars, 500 μm. (b) HOXA1 siRNAs disrupt normal cell cycle distribution. Top, histograms of Hoechst-33342 staining in human lung fibroblasts transfected with scrambled siRNAs and a HOXA1 siRNA pool 48 h after transfection (right). Bottom, same as above using HeLa cells 72 h after transfection. Cell cycle phase gates were drawn as approximations of the Dean-Jett-Fox cell cycle modeling algorithm. (c) HOXA1 siRNAs shift cell cycle distribution toward the G1 phase and sub-G1 phase fraction. Percentage of cells in each phase of the cell cycle (average three experiments) using gates from b with scrambled and HOXA1 siRNAs in human lung fibroblasts and in HeLa cells. (d) Representative dot plot of Annexin V (x-axis) versus propidium iodide (PI) (y-axis) analyses of scrambled (left) and HOXA1 siRNA (right) transfected lung fibroblasts (top) and HeLa cells (bottom). Cells populating the upper (late apoptosis) and lower (early apoptosis) right quadrants were designated as apoptotic cells. (e) Percentage of apoptotic cells (average of three experiments) populating upper and lower right quadrants from lung fibroblasts and HeLa cells transfected with scrambled or HOXA1 siRNAs as shown in d. Error bars indicate 95% confidence intervals in expression.
Discussion
Cuffdiff 2 performs differential analysis at transcript-level resolution of RNA-seq experiments and controls for both variability across replicates and uncertainty in abundance expression estimates caused by ambiguously mapped reads. In the absence of isoform switching or specialization, gene expression estimates reported by Cuffdiff 2 are consistent with those produced by count-based schemes. However, fold change in counts is a poor proxy for change in expression when there is substantial differential regulation of isoforms. Thus, although competing methods may offer higher gene-level sensitivity than Cuffdiff 2, they also report a higher background of false positives. In experiments where few genes are truly differentially expressed, this background could occlude the true positives. Cuffdiff 2 controls for cross-replicate variability and read-mapping ambiguity by using a model for fragment counts based on the beta negative binomial distribution. Experiments with real and simulated data show that Cuffdiff 2 is highly accurate at gene- and transcript-level resolution, even when used with benchtop sequencers.
Cuffdiff 2 performs integrated differential analysis of genes and transcripts within a single software workflow. Alternate means of performing differential analysis at transcript-level resolution that combine transcript-level fragment count estimates with existing count-based tools for assessing differential expression suffer from several limitations. Workflows combining methods fail to conform to several key requirements imposed by the component tools. For example, DESeq and edgeR expect that the input data are the number of perfectly and unambiguously mapped fragments that originate from each gene or transcript in each library. Failing to account for uncertainties in counts owing to ambiguous reads can result in false differential expression calls of transcripts with similar isoforms within the same gene, especially when sequencing depth is insufficient to accurately resolve the abundance of individual isoforms. Notably, our simulations show that this problem is more severe in genes with many isoforms, and cannot be eliminated by simply adding more replicates or sequencing depth to the experiment. Cuffdiff 2 surmounts this challenge by augmenting the cross-replicate variability modeling strategy used by count-based methods with incorporation of fragment assignment uncertainty computed for each gene. This enables it to dynamically control for uncertainty in highly complex or insufficiently sequenced genes. Recent large-scale transcriptome surveys have found that alternative splicing is extremely prevalent, with about three-quarters of human genes producing multiple abundant isoforms in a given cell type41. Moreover, thousands of human genes contain introns that have ‘NAGNAG’ splice sites, where N is any nucleotide and either AG can form an acceptor, generating isoforms that differ only by a single codon42. Thus, dealing with fragment assignment ambiguity is likely to be an increasingly important concern in differential analysis of RNA-seq data.
Commercially available library multiplexing kits have made sequencing-based designs cost-competitive with cDNA microarrays for expression analysis, but sequencing cost depends on overall sequencing depth, read length and number of replicates. Our simulations show that sensitivity is largely a function of depth, and specificity is mostly dependent on replication. However, long, paired reads dramatically aid in transcript and gene discovery, and we caution against the use of single reads in studies aimed at transcriptome assembly.
Cuffdiff 2 has offered a transcript-resolution view of the role of HOXA1, a critical regulator of embryonic development and body patterning, in maintaining adult cells. We have shown in different cell types that HOXA1 knockdown perturbs the expression of thousands of genes, alters the isoform selection of key cell cycle regulators and causes disruption of the cell cycle leading to cell death. Further experiments will be required to determine the nature and mechanism of the disruption and to identify the direct targets of HOXA1.
With Cuffdiff 2, RNA-seq can now be used for robust differential expression analysis at both gene- and isoform-level resolution with a single analysis tool. This creates opportunities for integrated genomic analysis of unprecedented scope and scale and can uncover biological phenomena not observable with other high-throughput technologies. Sequencing is now used to map histone modifications and protein-DNA interactions (ChIP-Seq43,44), chromatin accessibility (DNase hypersensitivity45, FAIRE46) and conformation (ChIA-PET47), and processing of RNA by protein (RIP-Seq48, CLIP-Seq49). Analyses with complementary sequencing assays are becoming increasingly common. For example, a recent study coupled transcript-level resolution RNA-seq with CLIP-Seq to track splicing changes that depend on muscleblind-like RNA binding proteins, which play key roles in development and in myotonic dystrophy50. The authors used high-throughput measurements of cellular state to connect sequence features of the genome to the transcriptional and post-transcriptional regulation of its genes. The large genomic footprints and numerous isoforms of many genes can greatly complicate such studies. Transcript-resolution measurements made with RNA-seq could drastically simplify the problem by eliminating unexpressed transcripts and isolating abundant ones. We are confident that the power and resolution offered by Cuffdiff 2 will allow biologists to better disentangle complex cellular circuitry and precisely relate genomic sequence to gene regulation.
Online Methods
Fold change measurement and differential analysis by raw count schemes
Raw counts for each gene in the UCSC coding gene annotation (hg19) were measured with HTSeq (http://www-huber.embl.de/users/anders/HTSeq/) version 0.5.3p3, using the “–stranded=no” option and either the “–mode=intersection-strict” option for exon-intersection counting or “–mode=union” for exon union counting. Differential expression was called using these counts with DESeq version 1.8.3 or edgeR version 2.6.8 according to the package vignettes and with an FDR of 1%. Fold changes in expression were reported by these packages after correcting for differences in library sizes.
Cell culture and siRNA knockdown
Embryonic lung fibroblasts cells (IMR-90) were obtained from ATCC (Cat#CCL-186) and grown in Dulbecco's modified Eagle's medium (DMEM, Invitrogen) with 10% FBS (Invitrogen) and supplemented with 100 U/ml penicillin, 100 mg/ml streptomycin, and 4 mM glutamine at 37 °C and 5% CO2. Reverse siRNA transfections were done using Lipofectamine RNAiMAX as per vendor's specifications (Invitrogen). Six-well dishes were plated at 80,000 (40–50% confluency) cells per well on the day of transfection with either HOXA1 siRNA pools (Dharmacon L-017464-00-0005) or nontargeting control pools (Dharmacon D-001810-10-20) at 25 nM using 5 μl of Lipofectamine reagent per well. HOXA1 knockdowns and nontargeting controls were preformed in triplicate. RNA was extracted 48 h after transfection using 1 ml or TriZol reagent (Invitrogen) and purified with RNeasy Mini kits. Genomic DNA was removed using on-column DNase I treatment according to the manufacturer's instructions (Qiagen).
Gene expression microarrays
For each knockdown/control 200 ng of total RNA was amplified and labeled with CY3 using the Agilent Low Input Quick Amp Labeling One Color Kits and hybridized to Agilent SurePrint G3 Gene Expression Microarrays as per manufacturer's specifications. Probe intensities were extracted using the Feature Extraction Software (GE1 Sep09 protocol).
mRNA-seq library preparation and sequencing
For each RNA sample, we prepared Illumina mRNA-seq libraries using the TruSeq RNA kit (version 1, rev A), using 1 μg of total RNA and prepared according to manufacturer's instruction. For HiSeq 2000 sequencing, eight libraries were pooled per sequencing lane (including libraries not described in this manuscript). One anti-HOXA1 siRNA library and one scrambled control library were pooled in each of three sequencing lanes, resulting in each of the six libraries discussed here being sequenced with ∼30 million reads. Human lung fibroblast reads are available at GEO accession GSE37704. ESC reads were downloaded from the ENCODE DCC hosted at the UCSC genome browser (Caltech RNA-seq section; http://genome.ucsc.edu/ENCODE/).
RNA-seq read alignment
RNA-seq reads for each library were mapped independently using TopHat version 1.4.0 (http://tophat.cbcb.umd.edu/) against the human genome build hg19, downloaded via the UCSC genome browser. HiSeq 2000 libraries were aligned with the following options: “-r 50 -p 8 –G ucsc_coding.gtf -a 5 -F 0.0,” where ucsc_coding.gtf contains the UCSC coding transcripts in GTF format. MiSeq libraries were mapped as above with the added option “—no-novel-juncs.”
Gene expression analysis by microarrays
The same RNA samples used as input for RNA-seq were also assayed by Agilent Microarrays. Normalization for the Agilent arrays was done by converting log2 intensity values to ratios of the 75th percentile intensity for each array after removal of control probes. Only probes passing the background threshold in more than 90% of the arrays were considered for further analysis. Remaining probes were collapsed around the median using the Agilent “GeneName” identifier (mixture of custom and HUGO gene symbol identifiers). Arrays were then batch corrected using the ComBat software package in R to correct for slide batch effects (eight arrays per slide). After all normalization, significant changes in gene expression between HOXA1 knockdowns and nontargeting controls were determined using Significance Analysis of Microarrays (SAM). Genes with a SAM-supplied q-value of ≤0.01 (1% FDR) were deemed significant. Microarray data are available at GEO accession GSE37704.
Gene and isoform expression analysis by RNA-seq
Gene and isoform expression levels were calculated as detailed in Supplementary Methods section 2 by running Cuffdiff 2 (http://cufflinks.cbcb.umd.edu/) on the alignments from TopHat and the UCSC coding genes. Briefly, library sizes were normalized using the method proposed by Anders and Huber17, with slight modifications described in Supplementary Methods section 2.1. Cross-replicate variability in gene-level fragment counts (overdispersion) was estimated by fitting a generalized linear model through the count variance as a function of the mean. Isoform abundances were calculated using the linear model described previously3,30, with sequence bias correction enabled and correcting for multiply mapping reads with the “-u” option of Cuffdiff 2. Fragment assignment uncertainty was calculated as described in Supplementary Methods section 2.4, resulting in a variance-covariance matrix ψ for each gene that captures the variability in assigning fragments to that gene's isoforms. This matrix is used in combination with the overdispersion model to parameterize a beta negative binomial distribution for the fragments generated by each isoform, as described in Supplementary Methods section 2.5. These distributions are used by Cuffdiff 2 to simulate sequencing of each isoform and subsequent assignment of fragments to the isoforms. From these simulations, Cuffdiff 2 derives variance-covariance matrices on assigned fragment counts, which allow it to calculate gene-level variance estimates as described in Supplementary Methods section 2.6. Testing for differential expression and shifts in relative isoform output are described in Supplementary Methods section 2.7.
Simulation of RNA-seq
RNA-seq simulations were performed with the TuxSim sequencing framework (http://cufflinks.cbcb.umd.edu/tuxsim/) and are described in detail in Supplementary Methods section 3. Briefly, TuxSim takes as input a file containing relative abundances for each transcript in a GTF file, and then generates reads from the transcripts according to their relative abundances. Paired-end reads were drawn uniformly at random from within transcripts, and follow a normal fragment length distribution (mean = 180 bp, s.d. = 50 bp). Alignments to the genome (hg19) for these reads were directly emitted as SAM file. The conditions compared in silico were based on the real sequencing data from the scrambled siRNA fibroblast control. The first condition modeled the unperturbed fibroblast data. Replicates were generated by selecting a fragment count for each transcript from a negative binomial variate generator, parameterized according to the fitted overdispersion in the real data. The second condition was sequenced similar to the first after 1,000 randomly selected genes had been perturbed in expression as described in Supplementary Methods section 3. The alignments for each condition were compared with Cuffdiff 2 using the same (default) options as used to process the real data.
Gene set enrichment analysis
Enrichment for up- or downregulation sets of genes from the REACTOME pathway database was computed by running GSEA against the fold-change ranked list of genes in the experiment. Ranking was based on Cuffdiff 2-derived fold change. REACTOME gene sets with between 12 and 200 members in the MSigDB package “c2.all.v3.0.symbols. gmt” were downloaded from ftp://gseaftp.broadinstitute.org/.
Cell cycle and apoptosis analyses with flow cytometry
For cell cycle analyses, lung fibroblasts and HeLa cells treated 48 h and 72 h, respectively, with either scrambled or HOXA1 siRNA pools were washed once with pre-warmed HBSS and incubated for 90 min at 37 °C in HBSS (Invitrogen, cat 14025092) + 2% FCS (FCS) + 10 mM Hepes pH 7.4 media containing 10 μg/mL of Hoechst-33342 (Invitrogen cat#H3570). After incubation, cells were trypsinized, washed twice with cold HBSS + 2% FCS (FCS) + 10 mM Hepes pH 7.4 media and resuspended in ∼200–500 μl cold HBSS + 2% FCS (FCS) + 10 mM Hepes pH 7.4 media in 5 ml round-bottom tubes for analysis. Flow cytometry analyses were done using an LSRII flow cytometer (Becton Dickinson).
For apoptosis analyses, lung fibroblasts and HeLa cells treated 48 h and 72 h, respectively, with either scrambled or HOXA1 siRNA pools were trypsinized, washed twice with cold PBS and resuspended at a concentration of 106 cells/ml in 1× binding buffer (10 mM HEPES pH7.4, 1 mM NaCl, 25 mM CaCl2). Cells were then aliquoted (100 μl) into 5 ml round-bottom tubes and incubated 15 min at room temperature with fluorescein isothiocyanate-labeled anti-Annexin V antibody (BD Biosciences cat#560931) and propidium iodide (Sigma P4864-10ML) as per manufacturer's instructions. Cell suspensions were brought up to 500 μl with 1× binding buffer and analyzed immediately on an LSRII flow cytometer (Becton Dickinson).
Supplementary Material
Acknowledgments
We are grateful to D. Kelley for a careful reading of the manuscript, and B. Wold for sharing the hESC RNA-seq data. We are also thankful for the ongoing development efforts of A. Roberts, B. Langmead, D. Kim, G. Pertea, H. Pimentel and S. Salzberg. C.T. and D.G.H. are Damon Runyon Postdoctoral Fellows. J.L.R. is a Damon Runyon-Rachleff Inovator fellow. This work was supported by US National Institutes of Health grants DP2OD006670, P01GM099117, P50HG006193 and RO1ES020260 (to J.L.R.) and R01 HG006129 and R01 DK094699 (to L.P.).
Footnotes
Accession code. GEO: GSE37704.
Note: Supplementary information is available in the online version of the paper.
Author Contributions: C.T. and L.P. developed the mathematics and statistics. D.G.H. and M.S. performed the experiments. D.G.H. and C.T. designed the experiments and performed the analysis. C.T. and L.G. implemented the software. L.P., J.L.R., D.G.H. and C.T. conceived the research. All authors wrote and approved the manuscript.
Competing Financial Interests: The authors declare no competing financial interests.
References
- 1.Cloonan N, et al. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat Methods. 2008;5:613–619. doi: 10.1038/nmeth.1223. [DOI] [PubMed] [Google Scholar]
- 2.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 3.Trapnell C, et al. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guttman M, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol. 2010;28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: An assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fu X, et al. Estimating accuracy of RNA-seq and microarrays with proteomics. BMC Genomics. 2009;10:161. doi: 10.1186/1471-2164-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Graveley BR, et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471:473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lister R, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pickrell JK, et al. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464:768–772. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Montgomery SB, et al. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature. 2010;464:773–777. doi: 10.1038/nature08903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang ET, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 13.Jiang H, Wong WH. Statistical inferences for isoform expression in RNA-seq. Bioinformatics. 2009;25:1026–1032. doi: 10.1093/bioinformatics/btp113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katz Y, Wang ET, Airoldi EM, Burge CB. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods. 2010;7:1009–1015. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nicolae M, Mangul S, Măndoiu II, Zelikovsky A. Estimation of alternative splicing isoform frequencies from RNA-seq data. Algorithms Mol Biol. 2011;6:9. doi: 10.1186/1748-7188-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee S, et al. Accurate quantification of transcriptome from RNA-seq data by effective length normalization. Nucleic Acids Res. 2011;39:e9. doi: 10.1093/nar/gkq1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Langmead B, Hansen KD, Leek JT. Cloud-scale RNA-sequencing differential expression analysis with Myrna. Genome Biol. 2010;11:R83. doi: 10.1186/gb-2010-11-8-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oshlack A, Robinson MD, Young MD. From RNA-seq reads to differential expression results. Genome Biol. 2010;11:220. doi: 10.1186/gb-2010-11-12-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang L, Feng Z, Wang X, Wang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26:136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- 22.Hardcastle TJ, Kelly KA. baySeq: empirical Bayesian methods for identifying differential expression in sequence count data. BMC Bioinformatics. 2010;11:422. doi: 10.1186/1471-2105-11-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Griffith M, et al. Alternative expression analysis by RNA sequencing. Nat Methods. 2010;7:843–847. doi: 10.1038/nmeth.1503. [DOI] [PubMed] [Google Scholar]
- 24.Glaus P, Honkela A, Rattray M. Identifying differentially expressed transcripts from RNA-seq data with biological variation. Bioinformatics. 2012;28:1721–1728. doi: 10.1093/bioinformatics/bts260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anders S, Reyes A, Huber W. Detecting differential usage of exons from RNA-seq data. Genome Res. 2012;22:2008–2017. doi: 10.1101/gr.133744.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pearson JC, Lemons D, McGinnis W. Modulating Hox gene functions during animal body patterning. Nat Rev Genet. 2005;6:893–904. doi: 10.1038/nrg1726. [DOI] [PubMed] [Google Scholar]
- 27.Xi W, WU Z, Zhang X. Isoform abundance inference provides a more accurate estimation of gene expression levels in RNA-seq. J Bioinform Comput Biol. 2010;08:177. doi: 10.1142/s0219720010005178. [DOI] [PubMed] [Google Scholar]
- 28.Tarazona S, García-Alcalde F, Dopazo J, Ferrer A, Conesa A. Differential expression in RNA-seq: a matter of depth. Genome Res. 2011;21:2213–2223. doi: 10.1101/gr.124321.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hiller D, Jiang H, Xu W, Wong WH. Identifiability of isoform deconvolution from junction arrays and RNA-seq. Bioinformatics. 2009;25:3056–3059. doi: 10.1093/bioinformatics/btp544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roberts A, Trapnell C, Donaghey J, Rinn JL, Pachter L. Improving RNA-seq expression estimates by correcting for fragment bias. Genome Biol. 2011;12:R22. doi: 10.1186/gb-2011-12-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rinn JL, Bondre C, Gladstone HB, Brown PO, Chang HY. Anatomic demarcation by positional variation in fibroblast gene expression programs. PLoS Genet. 2006;2:e119. doi: 10.1371/journal.pgen.0020119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu JQ, et al. Dynamic transcriptomes during neural differentiation of human embryonic stem cells revealed by short, long, and paired-end sequencing. Proc Natl Acad Sci USA. 2010;107:5254–5259. doi: 10.1073/pnas.0914114107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cabili MN, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011 doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morgan DO, Morgan DO. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu Rev Cell Dev Biol. 1997;13:261–291. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- 36.Liu S, et al. Structural analysis of human Orc6 protein reveals a homology with transcription factor TFIIB. Proc Natl Acad Sci USA. 2011;108:7373–7378. doi: 10.1073/pnas.1013676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dhar SK, Dhar SK. Identification and characterization of the human ORC6 homolog. J Biol Chem. 2000;275:34983–34988. doi: 10.1074/jbc.M006069200. [DOI] [PubMed] [Google Scholar]
- 38.Guillamot M, et al. Cdc14b regulates mammalian RNA polymerase II and represses cell cycle transcription. Scientific Reports. 2011;1:189. doi: 10.1038/srep00189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Washkowitz AJ, Gavrilov S, Begum S, Papaioannou VE. Diverse functional networks of Tbx3 in development and disease. Wiley Interdisciplinary Rev Syst Biol Med. 2012;4:273–283. doi: 10.1002/wsbm.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilson V, Wilson V, Conlon FL, Conlon FL. The T-box family. Genome Biol. 2002;3:S3008. doi: 10.1186/gb-2002-3-6-reviews3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Djebali S, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bradley RK, Merkin J, Lambert NJ, Burge CB. Alternative splicing of RNA triplets is often regulated and accelerates proteome evolution. PLoS Biol. 2012;10:e1001229. doi: 10.1371/journal.pbio.1001229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 44.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Crawford GE, et al. Genome-wide mapping of DNase hypersensitive sites using massively parallel signature sequencing (MPSS) Genome Res. 2006;16:123–131. doi: 10.1101/gr.4074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Giresi PG, Lieb JD. Isolation of active regulatory elements from eukaryotic chromatin using FAIRE (formaldehyde assisted isolation of regulatory elements) Methods. 2009;48:233–239. doi: 10.1016/j.ymeth.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fullwood MJ, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao J, et al. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Licatalosi DD, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang ET, et al. Transcriptome-wide regulation of pre-mRNA splicing and mRNA localization by muscleblind proteins. Cell. 2012;150:710–724. doi: 10.1016/j.cell.2012.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.