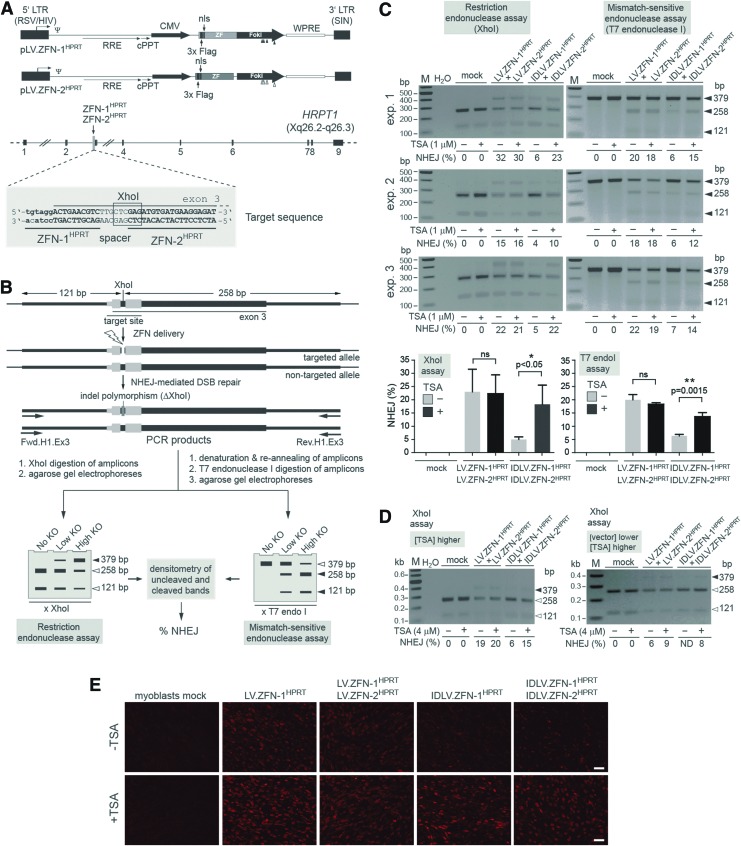
FIG. 1.
Effect of HDAC inhibition on target gene disruption in human myoblasts after IDLV- versus LV-mediated ZFN delivery. (A) Upper panel: genetic composition of the LV transfer constructs coding for the HPRT1-specific ZFNs. The plasmids pLV.ZFN-1HPRT and pLV.ZFN-2HPRT encode the custom-made nucleases ZFN-1HPRT and ZFN-2HPRT. Their DNA-binding zinc-finger arrays are directed at the upstream end of exon 3 of HPRT1. Close to their N-termini, the ZFNs harbor Flag epitopes and the SV40 large T antigen's nls. Boxes with broken arrow, hybrid 5′LTR consisting of Rous sarcoma virus- and HIV-1-derived sequences; boxes without broken arrow, SIN 3′LTR; ZF, nucleotide sequences coding for the zinc-finger arrays; FokI, sequences encoding mutant FokI nuclease motifs, with the positions of the nucleotide changes engineered to code for residues EELI and QKIK being indicated by solid and open vertical arrowheads, respectively. These FokI variants form obligate target site-specific heterodimers resulting in reduced ZFN-associated cytotoxicity. Ψ, HIV-1 packaging signal; RRE, Rev-responsive element; cPPT, central polypurine tract; WPRE, Woodchuck hepatitis virus posttranscriptional regulatory element. The ZFN ORFs are under the transcriptional control of the CMV immediate-early gene promoter and the polyadenylation signal located within the SIN 3′LTR. The plasmid backbone containing the prokaryotic origin of replication and the β-lactamase gene are not drawn. Lower panel: HPRT1 target locus. The nine exons of this X-linked gene are indicated as solid boxes, and the location of the ZFN-1HPRT and ZFN-2HPRT target sites is pinpointed by the vertical arrow. The HPRT1 segment corresponding to exon 3 is shown in upper case, while that of its contiguous upstream intron sequence is depicted in lower case. The 6 bp sequence flanked by the two ZFN target sites (spacer) partially overlaps with an XhoI recognition site (boxed nucleotides). (B) Schematic representation of the XhoI- and T7 endonuclease I-based genotyping assays employed to measure targeted DSB formation. DNA breaks at the HPRT1 target site can be repaired via the error-prone NHEJ pathway resulting in the introduction of polymorphisms in the form of indels that, in effect, can KO the XhoI recognition sequence. PCR on cellular DNA isolated from cells exposed to ZFN-1HPRT and ZFN-2HPRT should yield amplicons corresponding to targeted and nontargeted alleles. After electrophoresis of XhoI-treated amplicons, the frequencies of DSBs in different target cell populations (i.e., % NHEJ) can be determined through densitometry of XhoI-resistant and XhoI-digested DNA bands (left-hand diagram, solid and open arrowheads, respectively). In parallel or in alternative, amplicons corresponding to targeted and nontargeted alleles can be subjected to a cycle of denaturation and re-annealing of their DNA strands resulting in wild type: mutated heteroduplexes with mismatched base pairs. The T7 endonuclease I by cleaving at DNA sequences with such mismatched based pairs yields low-molecular-weight DNA fragments. After electrophoresis of T7 endonuclease I-treated amplicons, the rate of DSB formation in different target cell populations (i.e., % NHEJ) can be determined through densitometry of full-length and cleaved PCR products (right-hand diagram, open and solid arrowheads, respectively). (C) Analyses of XhoI- and T7 endonuclease I-treated PCR products amplified from genomic DNA of myoblasts transduced with the vectors encoding HPRT1-specific ZFNs. Myoblasts were incubated with 1:1 mixtures of LV.ZFN-1HPRT and LV.ZFN-2HPRT or of IDLV.ZFN-1HPRT and IDLV.ZFN-2HPRT. The total vector dose applied was 3.6×107 vgc/ml. During the 16 hr vector transduction period, the myoblast cultures were not exposed (−) or were exposed (+) to 1 μM of TSA. Mock-transduced cells served as negative controls. The genomic DNA was isolated at 48 hr posttransduction, and target gene disruption assays were deployed as outlined in Fig. 1B. The plots in the lower part of the panel correspond to the mean±standard deviation from three independent experiments examined by using XhoI- and T7 endonuclease I-based readouts. ns, not significant. (D) Analyses of XhoI-treated PCR products amplified from genomic DNA of myoblasts transduced with the vectors encoding HPRT1-specific ZFNs. Myoblasts were incubated with 1:1 mixtures of LV.ZFN-1HPRT and LV.ZFN-2HPRT or of IDLV.ZFN-1HPRT and IDLV.ZFN-2HPRT. The total vector doses applied were 3.6×107 vgc/ml (left panel) and 2.4×107 vgc/ml (right panel). During the 16 hr vector incubation period, the myoblast cultures were not exposed (−) or were exposed (+) to 4 μM of TSA. The genomic DNA was extracted at 48 hr posttransduction to determine XhoI site disruptions. ND, not detected. (E) Immunofluorescence microscopy analysis of ZFN expression in untreated and TSA-treated myoblasts. Target cells were mock-transduced or were transduced with LV.ZFN-1HPRT and IDLV.ZFN-1HPRT at 1.8×107 vgc/ml each or were cotransduced with 1:1 mixtures consisting of LV.ZFN-1HPRT and LV.ZFN-2HPRT or of IDLV.ZFN-1HPRT and IDLV.ZFN-2HPRT at a total vector dose of 3.6×107 vgc/ml. ZFN synthesis was accessed by Flag tag-specific immunofluorescence microscopy at 48 hr posttransduction. Size bars correspond to 200 μm. CMV, cytomegalovirus promoter; DSB, double-stranded DNA break; HDAC, histone deacetylase; IDLV, integrase-defective lentiviral vector; KO, knockout; NHEJ, nonhomologous end-joining; nls, nuclear localization signal; ORFs, open reading frames; PCR, polymerase chain reaction; SIN, self-inactivating; TSA, trichostatin A; ZFN, zinc-finger nuclease. Color images available online at www.liebertpub.com/hgtb