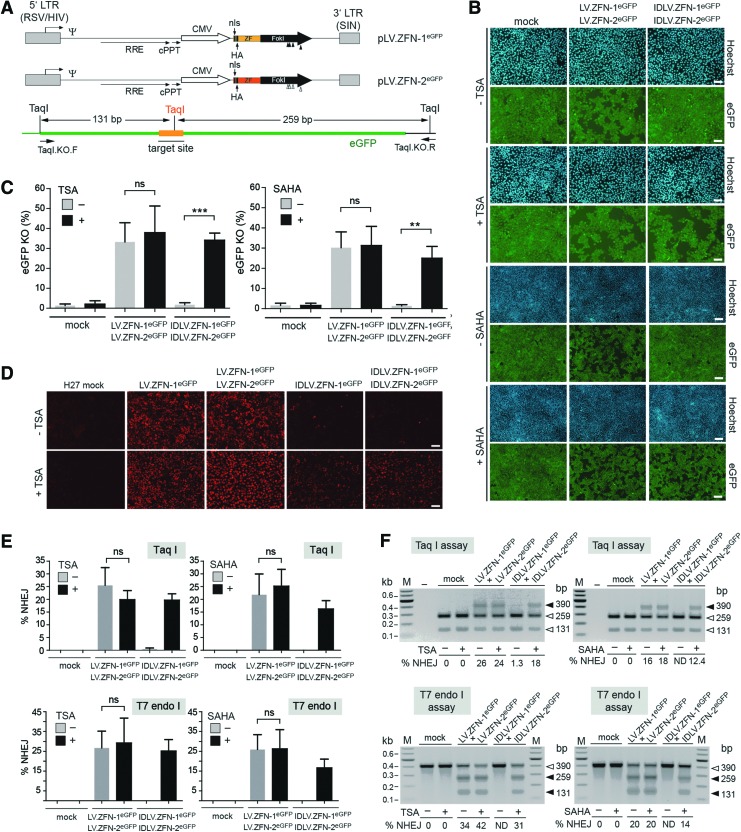
FIG. 4.
TSA- and SAHA-dependent rescue of target gene knockout levels in reporter H27 cells after IDLV-mediated ZFN delivery. (A) Upper panel: genetic organization of the LV transfer constructs coding for the eGFP-specific ZFNs. The plasmids pLV.ZFN-1eGFP and pLV.ZFN-2eGFP code for the sequence-specific nucleases ZFN-1eGFP and ZFN-2eGFP. The ZFNs contain nls and influenza HA epitopes located close to their N-termini. The FokI positions engineered to encode the heterodimer-forming residues EEAI and QKIV are indicated by solid and open vertical arrowheads, respectively. For a description of regulatory sequences and viral cis-acting elements, see the legend of Fig. 1A. Lower panel: schematic representation of the recombinant chromosomal region targeted by the eGFP-specific ZFNs. TaqI recognition sequences flanking and at the hybrid nucleases' target site (orange line) are indicated together with the expected TaqI DNA fragment sizes resulting from unmodified alleles. The primer set used to amplify this region is also shown. (B) Fluorescence microscopy on monolayers of eGFP-expressing H27 cells. The reporter cells treated or not treated with TSA or SAHA were mock-transduced or were cotransduced with 1:1 mixtures of LV.ZFN-1eGFP and LV.ZFN-2eGFP or of IDLV.ZFN-1eGFP and IDLV.ZFN-2eGFP at a total vector dose of 6×106 vgc/ml. H27 cell nuclei in each microscopic field were identified through Hoechst 33342 staining, whereas target gene expression was monitored via eGFP-directed fluorescence microscopy at 10 days posttransduction. Note the dissimilar amounts of eGFP-negative cells. Size bars in micrographs corresponding to TSA- and SAHA-treated H27 cultures are equivalent to 200 and 500 μm, respectively. (C) Flow cytometry-based quantification of eGFP knockout levels in H27 cultures subjected to the aforementioned experimental conditions. The flow cytometric analyses were carried out at 10 days posttransduction. Plots correspond to mean±standard deviation from three independent experiments (D) Immunofluorescence microscopy analysis of ZFN expression in untreated and TSA-treated H27 cells. Target cells were mock-transduced or were transduced with LV.ZFN-1eGFP and IDLV.ZFN-1eGFP at 3×106 vgc/ml each or were cotransduced with 1:1 mixtures consisting of LV.ZFN-1eGFP and LV.ZFN-2eGFP or of IDLV.ZFN-1eGFP and IDLV.ZFN-2eGFP at a total vector dose of 6×106 vgc/ml. ZFN synthesis was accessed by HA-specific immunofluorescence microscopy at 48 hr posttransduction. Size bars correspond to 200 μm. (E) Cumulative results derived from three independent experiments performed on H27 indicator cells deploying the aforementioned experimental conditions. Data shown correspond to mean±standard deviation (F) Representative agarose gel electrophoresis of TaqI- and T7 endonuclease I-treated PCR products amplified from genomic DNA of H27 cells subjected to the aforementioned experimental conditions. HA, hemagglutinin; ND, not detected; vgc, vector genome copies. Color images available online at www.liebertpub.com/hgtb