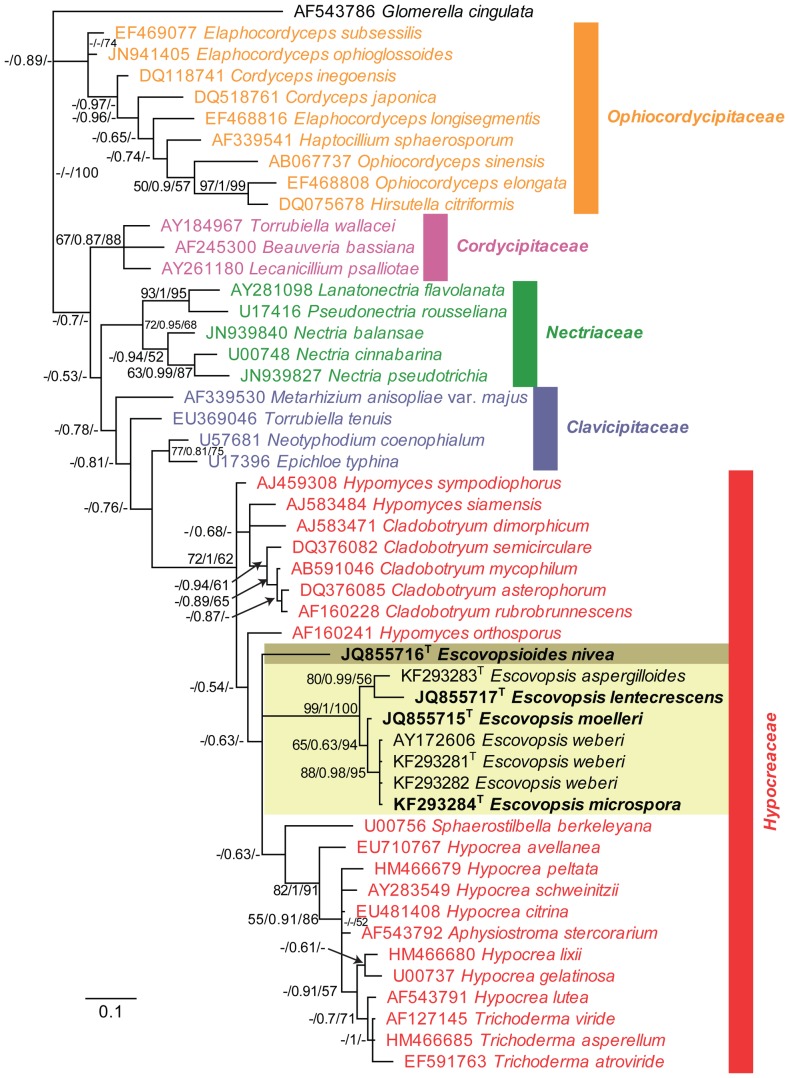
Figure 8. Fifty-percent majority rule tree obtained from the Bayesian analysis of LSU DNA sequence data.
The Escovopsis and Escovopsioides clades are highlighted in shaded boxes. In these clades, a superscript T denotes sequences obtained from (ex-) type strains. Regular type taxa in the clade ascribe sequences obtained from GenBank, which were included for the purpose of phylogenetically placing the new species. Except for some rearrangements at the deeper nodes (see TreeBASE), this Bayesian consensus tree is topologically identical to trees obtained from maximum parsimony (MP) and maximum likelihood (ML) analyses. Numbers at tree nodes are Maximum likelihood bootstrap support values (integer value up to 100), posterior probabilities support values obtained from Bayesian analysis (fraction up to 1) and parsimony bootstrap support values (integer value up to 100), respectively. Colour taxa indicate representatives from the hypocrealean families shown on the tree. The scale bar represents the expected number of changes per site. Glomerella cingulata was used to root the tree.