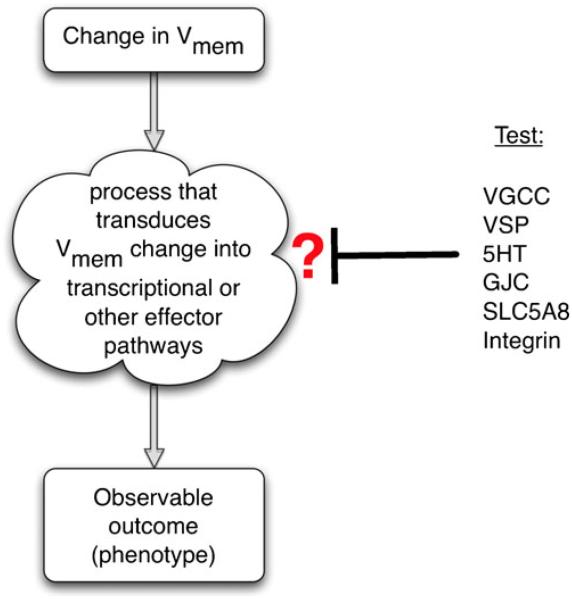
Fig. 3.
Identification of a transduction mechanism. To identify which of several possible mechanisms is involved in transducing an epigenetic (biophysical) event into a biochemical signaling pathway, a suppression screen should be performed (Blackiston et al. 2011). Each candidate transduction mechanism can be probed in turn by inhibiting it in an assay in which an induced change in transmembrane potential leads to an observable phenotype (increase in proliferation, up-regulation of a target gene, cell migration, etc.). For example, if gene X is up-regulated upon depolarization but this up-regulation does not occur in the presence of a GdCl, then voltage-gated calcium channel (VGCC) opening is implicated. If on the other hand a blocker of gap-junctional communication (GJC) abolishes the effects of depolarization, this suggests that the voltage-gated control of GJC and small molecule movement mediate the effects. Similar experiments with inhibitory drugs, dominant negative constructs and genetic deletion can be used to implicate serotonergic (5HT) signaling, butyrate-driven chromatin modifications (SLC5A8), integrin signaling, voltage-sensitive phosphatases (VSP), or other transduction mechanisms