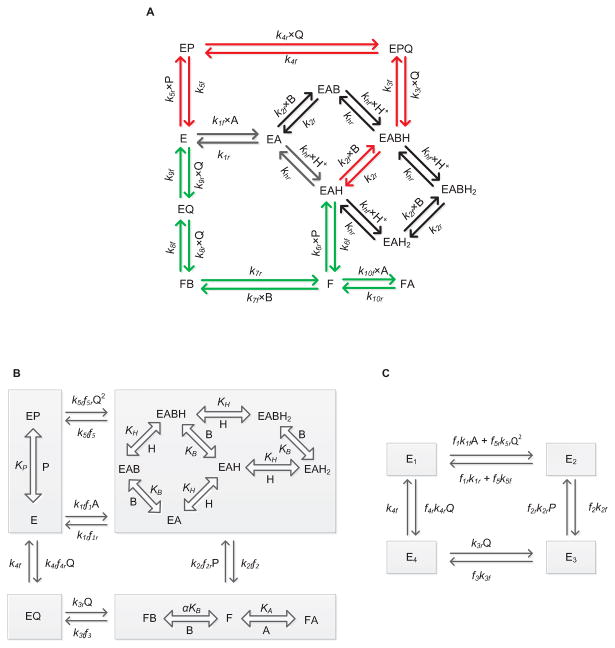
Figure 1.
(A) Basic scheme for the catalytic mechanism of glutathione reductase (GR). Free enzyme (E) interacts with the substrate NADPH first and random protonation and GSSG binding takes place at this complex to form activated single proton bound intermediate complex (E-NADPH-H) and dead-end complexes. Branching takes place at this complex (E-NADPH-H) to form sequential (red) and ping-pong branches (green). E and F indicate oxidized and reduced forms of the free enzyme, respectively. EX and EXH (X = NADPH (A), GSSG (B), NADP+ (P) and GSH (Q)) represents enzyme substrate/product complexes. kif and kir (i = 1-10) are forward and backward rate constants for the respective interaction. khf and khr are forward and backward rate constants for the proton binding to the intermediate complex and formation of dead-end complexes. (B, C) Reduced scheme of GR under rapid-equilibrium assumption. A, B, P, Q and H represent concentrations of NADPH, GSSG, NADP+, GSH and protons, respectively. KB, KP KH and KIA are dissociation constants for the respective rapid equilibrium interactions and α is the cooperative binding for GSSG. kif and kir (i = 1-5) are forward and backward rate constants for the respective interactions. E1, E2, E3 and E4 are intermediate enzyme states and fi indicates the associated binding polynomial for each enzyme-complex transition.