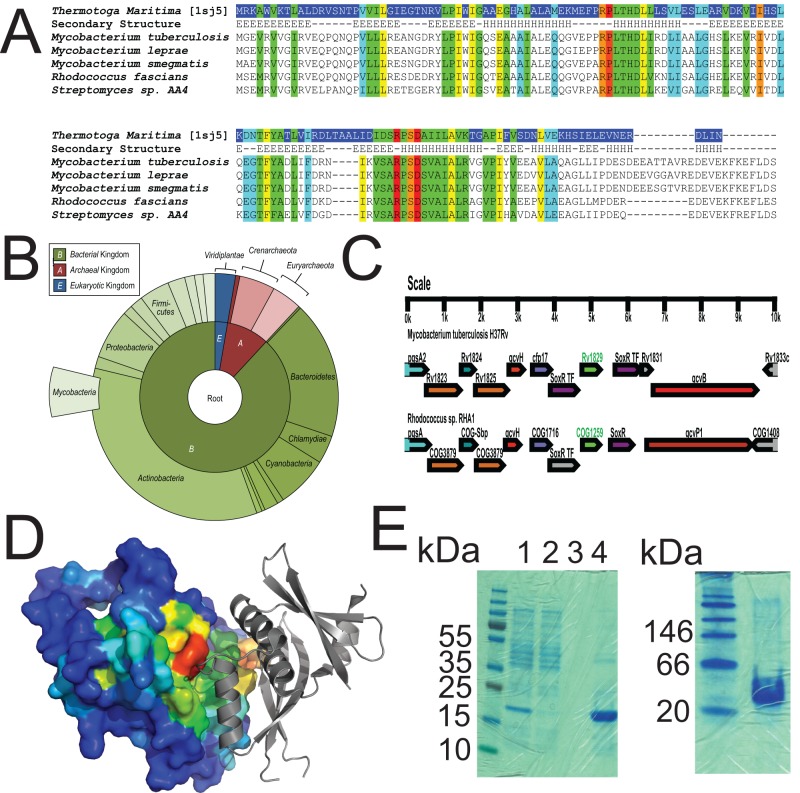
FIG 2 .
Cor is a conserved, ancient protein. (A) Alignment of Rv1829 from M. tuberculosis to orthologues from Thermotoga maritima, M. leprae, M. smegmatis, Rhodococcus fascians, and Streptomyces species AA4. Sequence alignment of Rv1829 homologues shows conserved amino acids (in rainbow colors) by conservation, with invariant nonhydrophobic residues colored in red. This color scheme is identical in the modeled crystal structure in panel D. (B) Species-distribution taxonomic tree in “sunburst” format. The distribution and evolutionary conservation of all known homologues of Cor are shown. (C) Alignment of the Cor genomic region from M. tuberculosis with the orthologous genomic region from Rhodococcus sp. demonstrates conservation of multiple surrounding genes. (D) The Cor sequence was mapped to the representative DUF151 structure, which highlighted a conserved surface cleft formed at the interface (also relatively conserved) of two DUF151 monomers (shown in red). (E) Cor was purified from E. coli as a 6×His-tagged protein and then run on a denaturing SDS-PAGE gel (left panel) or a native gel (right panel) with appropriate molecular mass markers. The predicted molecular masses of cor are 18 kDa for the monomer and 36 kDa for the dimer. In the left panel, lanes are total lysate (lane 1), flowthrough (lane 2), final wash (lane 3), and imidazole elution (lane 4).