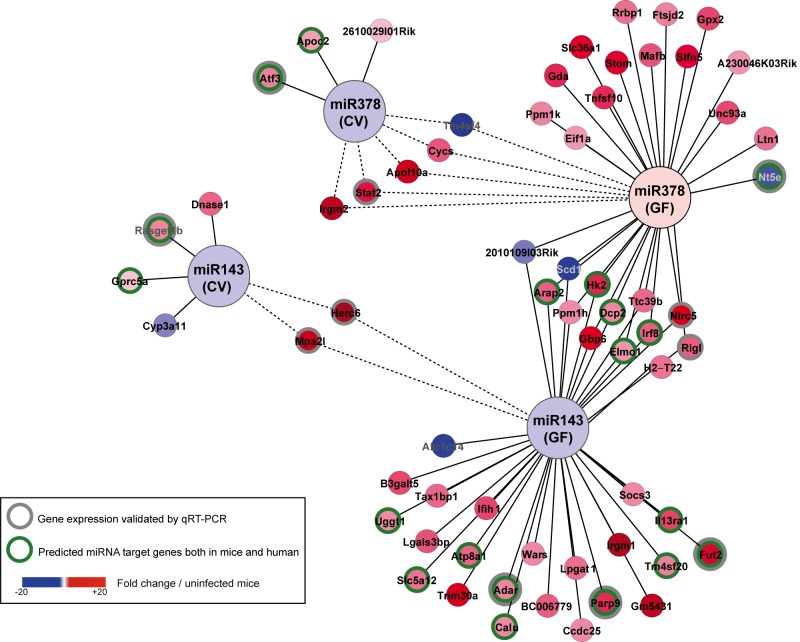
FIG 4 .
miRNA-mRNA network. Using Cytoscape, we highlighted, among the 5 miRNAs whose expression was affected by L. monocytogenes and the intestinal microbiota, a network linking miR-143 and miR-378 and the regulated host genes in the small intestine of conventional (CV) and germfree (GF) mice orally infected with L. monocytogenes 72 h p.i. As described in the text, miR-143 decreased upon infection in both CV and GF mice; in contrast, miR-378 decreased in CV mice but not in GF mice. Nodes represent infection-regulated miRNAs and genes; lines link each miRNA to its putative targets. Some of the predictions in mice were valid in humans (green circles). The color coding indicates the fold change of gene expression in Listeria-infected mice relative to the expression level in uninfected control mice. The differential expression levels of 11 genes were validated by RT-qPCR, as shown in Fig. 3B. Dotted lines show a gene that is similarly regulated in CV and GF mice.