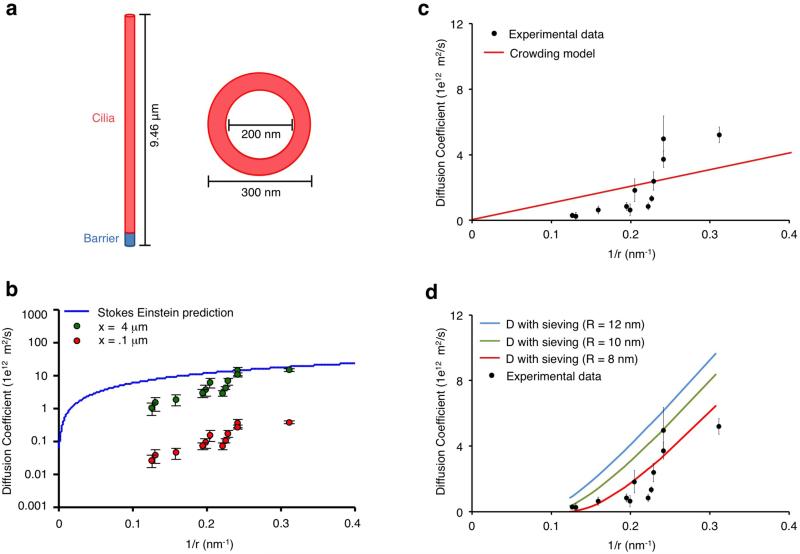
Figure 4.
Analysis of the diffusion barrier at primary cilia. (a) Structural depiction of the geometry of the primary cilia used to calculate diffusion coefficients. (b) Variation of barrier length and its effect on diffusion coefficients. Increases in the barrier length cause an upward shift in diffusion coefficients. The Stokes Einstein equation curve is included as an upper boundary to the diffusion coefficients. (c) Experimentally derived diffusion coefficients are plotted as a function of the inverse of their Stokes radii in comparison to a molecular crowding model shown in red. (d) Experimentally derived diffusion coefficients are plotted as a function of the inverse of their Stokes radii in comparison to a molecular sieving model. The resulting curves for different mesh sizes are included in the plot with an optimal fit at R = 8 nm. The YFP-FKBP probe proteins plotted are the followings: (in order of size): YFP-FKBP, YFP-FKBP-PKIM, YFP-FKBP-Grp1(229-772), YFP-FKBP-Grp1(229-1200), YFP-Luciferase-FKBP, YFP-FKBP-Luciferase, YFP-FKBP-Grp1(1-772), YFP-FKBP-Grp1(1-1200), YFP-FKBP-Tiam1, YFP-FKBP-ΔN β-Gal, YFP-β-Gal-FKBP, and YFP-FKBP-β-Gal.