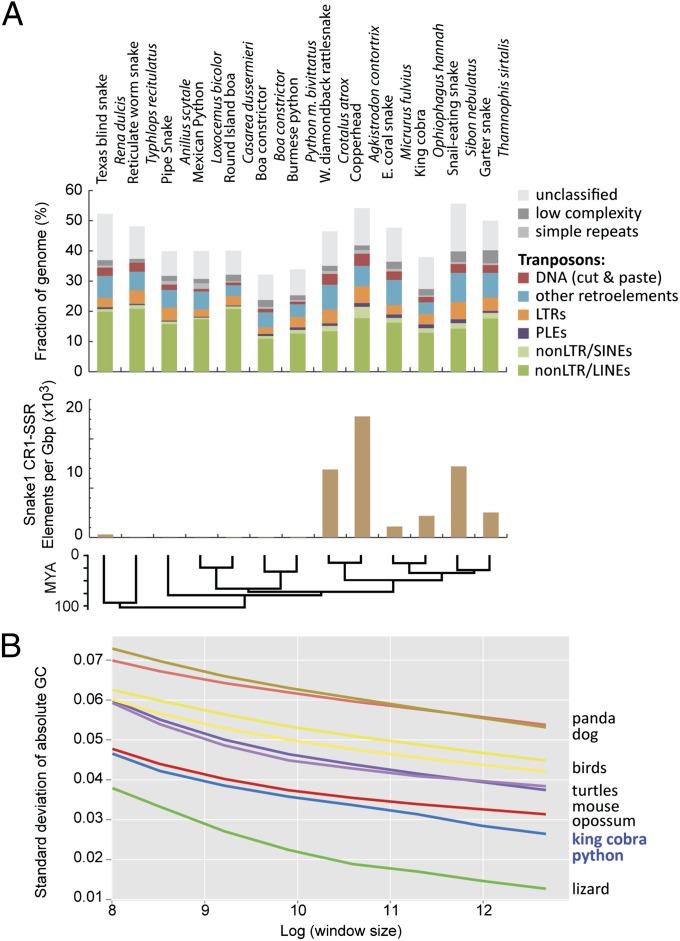
Fig. 4.
Variation and uniqueness of snake genome content and structure. (A) Amounts and types of readily identified repeat elements in snake complete and sampled genomes. Estimates in Top and Middle show abundance of genomic repeat elements across 10 snake species based on sampled genome sequences, except for the python and cobra, which are based on complete genomes. Bottom shows genomic density of snake1 CR1 LINE elements for subfamilies that tend to contain microsatellite repeats at their 3′ tails in snake genomes and genome samples. (B) Evidence for shifts in genomic GC isochore structure in squamate reptiles. The y axis is the standard variation of GC content when examining the genome at nonoverlapping window sizes (from 5- to 320-kb windows log-transformed on the x axis). Larger values indicate greater among-window GC content heterogeneity. For example, at all spatial scales, mammals have the greatest GC heterogeneity, and squamate reptiles have the least GC heterogeneity. The right side of the graph (GC SD at a window size of 320 kb) shows GC heterogeneity at a spatial scale on the order of isochore structure; the low GC heterogeneity of squamate reptiles indicates a reduced representation of GC-rich isochores compared with the other taxa. LTR, long terminal repeat; PLE, Penelope-like element; SINE, short interspersed nuclear element.