Significance
The ability to repair breaks in the DNA phosphodiester backbone is essential for genome integrity. When breakage results in 5′-PO4 and 3′-OH termini, the ends can be rejoined to each other, or to novel partner strands, by classic DNA ligases that covalently activate the 5′-PO4 end by linkage to AMP. However, when breakage leaves 5′-OH and 3′-PO4 termini, the ends are considered “dirty” because they cannot be sealed by classic ligases. This paper shows that the unconventional ligase RtcB can evade the DNA dirty end chemistry problem by splicing DNA 3′-PO4 ends to DNA 5′-OH ends. RtcB accomplishes this by attaching a GMP nucleotide to the DNA 3′-PO4 end to activate it for nucleophilic attack by the 5′-OH.
Abstract
There are many biological contexts in which DNA damage generates “dirty” breaks with 3′-PO4 (or cyclic-PO4) and 5′-OH ends that cannot be sealed by DNA ligases. Here we show that the Escherichia coli RNA ligase RtcB can splice these dirty DNA ends via a unique chemical mechanism. RtcB transfers GMP from a covalent RtcB–GMP intermediate to a DNA 3′-PO4 to form a “capped” 3′ end structure, DNA3′pp5′G. When a suitable DNA 5′-OH end is available, RtcB catalyzes attack of the 5′-OH on DNA3′pp5′G to form a 3′–5′ phosphodiester splice junction. Our findings unveil an enzymatic capacity for DNA 3′ capping and the sealing of DNA breaks with 3′-PO4 and 5′-OH termini, with implications for DNA repair and DNA rearrangements.
The Escherichia coli RtcB is a founding member of a recently discovered family of RNA repair/splicing enzymes that join RNA 2′,3′-cyclic-PO4 or 3′-PO4 ends to RNA 5′-OH ends (1–4). RtcB executes a four-step pathway that requires GTP as an energy source and Mn2+ as a cofactor (5–7). RtcB first reacts with GTP to form a covalent RtcB-(histidinyl337-N)–GMP intermediate. It then hydrolyzes the RNA 2′,3′-cyclic-PO4 end to a 3′-PO4 and transfers guanylate from His337 to the RNA 3′-PO4 to form an RNA3′pp5′G intermediate. Finally, RtcB catalyzes the attack of an RNA 5′-OH on the RNA3′pp5′G end to form the 3′–5′ phosphodiester splice junction and liberate GMP.
The unique chemical mechanism of RtcB overturned a long-standing tenet of nucleic acid enzymology, which held that synthesis of polynucleotide 3′–5′ phosphodiesters proceeds via the attack of a 3′-OH on a high-energy 5′-phosphoanhydride: either a nucleoside 5′-triphosphate in the case of RNA/DNA polymerases or an adenylylated intermediate A5′pp5′N, in the case of classic RNA/DNA ligases. In light of the wide distribution of RtcB proteins in bacteria, archaea, and metazoa, we raised the prospect of an alternative enzymology based on covalently activated 3′-PO4 ends (6).
In principle, the chemistry of RNA 3′-PO4/5′-OH end joining by RtcB might be portable to DNA transactions and pertinent to DNA repair. A variety of hydrolytic nucleases incise the DNA phosphodiester backbone to yield 3′-PO4 and 5′-OH termini that cannot be joined by DNA ligases. Nonligatable 3′-PO4 ends are also generated during base excision repair catalyzed by DNA glycosylase/lyase enzymes, during the repair of trapped covalent topoisomerase IB–DNA adducts by tyrosyl-DNA phosphodiesterase 1, and during DNA damage inflicted by ionizing radiation. One way nature solves this “dirty end” problem is by deploying a variety of “end healing” enzymes (8–14). These include 3′-phosphoesterases that convert a 3′-PO4 to a 3′-OH and 5′-kinases that transform a 5′-OH to a 5′-PO4, thereby enabling break sealing by the classic ligase pathway. Given what we now know about RtcB, would it not make sense for nature to also endow a pathway for direct joining of DNA 3′-PO4 and 5′-OH ends, be it via RtcB or another ligase yet to be discovered?
We can extend this thought to DNA breaks with 2′,3′-cyclic-PO4 and 5′-OH ends, which are generated by topoisomerase IB transesterification at sites where single ribonucleotides are embedded in the genome (15). Embedded ribonucleotides are among the most abundant natural “lesions” in DNA and their cleavage by topoisomerase IB causes a sharp increase in deletion mutations at the incision site (16, 17). Although the metabolism of 2′,3′-cyclic-PO4 DNA breaks is entirely uncharted, such ends are potential fodder for the cyclic phosphodiesterase activity of RtcB and subsequent end joining to restore the DNA backbone. If RtcB seals the duplex nick without intervening processing steps, the effect of such a faithful ligation could be to redirect repair of the embedded ribonucleotide to an apparently “safer” pathway driven by RNase H2 (18, 19). Alternatively, RtcB might splice the 2′,3′-cyclic-PO4 DNA end to a different 5′-OH acceptor, resulting in a mutagenic repair outcome. Single embedded ribonucleotides in DNA are also susceptible to incision by transesterifying ribonucleases that would generate DNA-monoribonucleoside-3′-PO4 and 5′-OH DNA ends; these breaks are potential substrates for ligation via RtcB-like chemistry. Here we interrogated the capacity of RtcB to splice DNA ends.
Results and Discussion
RtcB Splices DNA with a Ribonucleoside 3′-PO4 End.
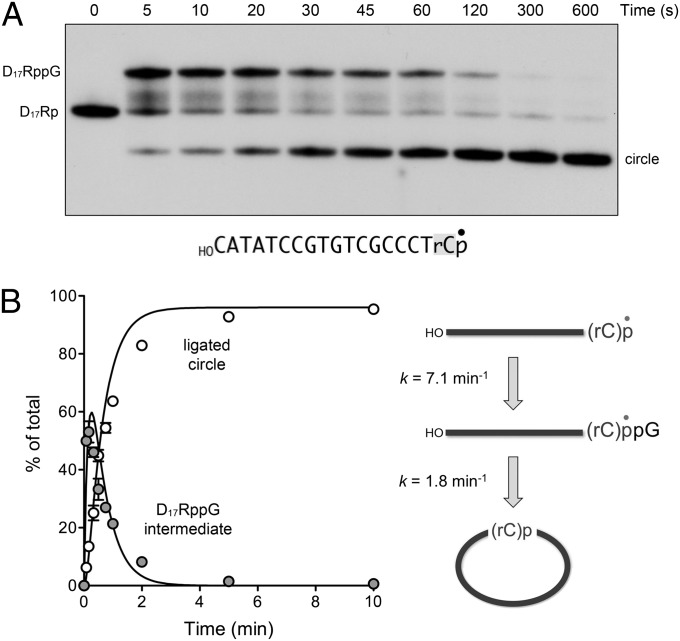
Initial experiments were performed using a HOD17Rp substrate with 5′-OH and 3′-PO4 termini that consisted of 17 deoxynucleotides and a single 32P-labeled ribonucleoside 3′-PO4. We found that RtcB, under single-turnover conditions in the presence of GTP and Mn2+, rapidly and efficiently ligated the 5′-OH and 3′-PO4 ends of HOD17Rp to form a circular product (Fig. 1A). A transient HOD17RppG intermediate (Fig. 1A) accumulated to an extent of 50% and 53% of the total nucleic acid at the 5- and 10-s time points and decayed steadily thereafter (Fig. 1B). From the kinetic profile, we derived rate constants of 7.1 ± 0.6 min−1 for the conversion of HOD17Rp to HOD17RppG and 1.8 ± 0.09 min−1 for the intramolecular splicing of HOD17RppG to form a circle (Fig. 1B). RtcB also catalyzed intramolecular splicing of a HOD17R>p substrate with a 2′,3′-cyclic-PO4 end (Fig. S1), a finding consistent with the fact that RtcB has an intrinsic ability to hydrolyze a 2′,3′-cyclic phosphodiester to a 3′-phosphomonoester (5, 7). Here, too, the HOD17RppG intermediate accumulated at the 5- and 10-s time points (comprising 44% and 37% of the total nucleic acid) and declined thereafter as the circular product was formed (Fig. S1).
Fig. 1.
RtcB efficiently splices 5′-OH DNA with a ribonucleoside-3′-PO4 terminus. (A) Reaction mixture containing 50 mM Tris⋅HCl (pH 8.0), 2 mM MnCl2, 100 µM GTP, 1 µM RtcB, and 0.02 µM HOD17Rp (shown with the 3′ ribonucleoside shaded and the 32P-label denoted by •) was incubated at 37 °C. Aliquots were withdrawn at the times specified and quenched immediately. The products were analyzed by urea-PAGE and visualized by autoradiography. The positions of the linear substrate and guanylylated intermediate are indicated (Left); the position of the ligated circle is indicated (Right). (B) The kinetic profile of the intramolecular end joining reaction is plotted. Each datum in the graph is the average of three separate experiments ±SEM. The data were fit by nonlinear regression in Prism to a unidirectional two-step pathway of 3′-PO4 guanylylation and phosphodiester synthesis (Right), with rate constants as indicated.
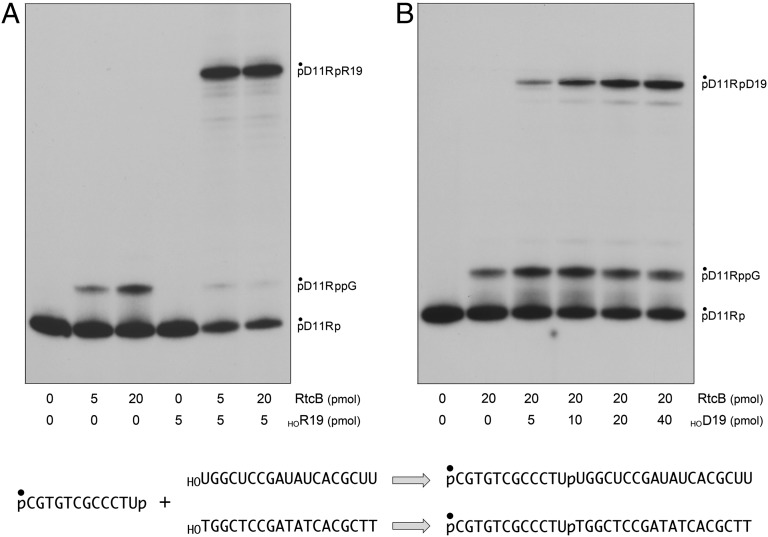
To query whether RtcB could also perform intermolecular splicing of a DNA 5′-OH end, we exploited a pD11Rp substrate composed of 11 deoxynucleotides and a single ribonucleoside 3′-PO4. The installation of a 5′-PO4 end on pD11Rp precludes circularization, but permitted guanylylation of the 3′-PO4 to generate pD11RppG (Fig. 2A). Intermolecular sealing was assayed by including a second unlabeled acceptor strand with 5′-OH and 3′-OH ends. The acceptor strands were all-RNA or all-DNA 19-mer oligonucleotides, of otherwise identical nucleobase sequence, that had no base complementarity to the pD11Rp donor strand (Fig. 2). A positive control showed that RtcB ligated pD11Rp to the HORNA acceptor to form pD11RpR19 (Fig. 2A). RtcB also joined pD11Rp to the HODNA acceptor, albeit less efficiently (8% ligation) than to HORNA (68% ligation) when the reaction mixtures contained 20 pmol (2 µM) enzyme and 5 pmol (0.5 µM) acceptor (Fig. 2B). The yield of intermolecular splicing to a HODNA acceptor rose to 38% when the acceptor level was increased to 40 pmol (4 µM) (Fig. 2B), suggesting that RtcB was less avid in capturing in trans a 5′-OH DNA end versus an RNA end. Collectively, these experiments demonstrate that RtcB can ligate 3′-PO4 and 5′-OH DNA ends punctuated by a single ribonucleoside 3′-PO4 (or 2′,3′-cyclic-PO4). Therefore, RtcB enzymes are plausible candidates to join ends generated by hydrolysis or transesterification of DNA at sites where single ribonucleotides are embedded.
Fig. 2.
Intermolecular splicing of ribonucleoside-3′-PO4 terminated DNA. Reaction mixtures (10 µL) containing 50 mM Tris⋅HCl (pH 8.0), 2 mM MnCl2, 100 µM GTP, 0.5 pmol (0.05 µM) of 5′ 32P-labeled pD11Rp substrate, 0, 5, or 20 pmol (0, 0.5, or 2 µM) of RtcB as indicated, and either an unlabeled 5′-OH 19-mer RNA acceptor strand (A) or DNA acceptor strand (B) as specified were incubated at 37 °C for 30 min. The products were analyzed by urea-PAGE and visualized by autoradiography. The positions of the substrate, guanylylated intermediate, and ligated product are indicated (Right). The substrate, acceptors, and reaction products are shown (Lower) with the 5′ 32P-label denoted by •.
Capping and Splicing of DNA 3′-PO4 Ends.
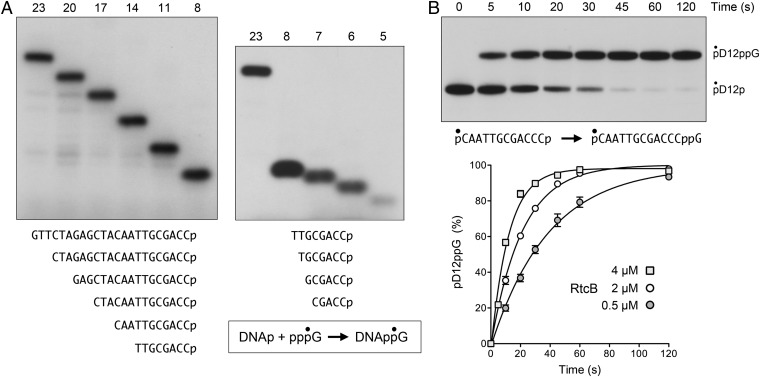
The next series of experiments was designed to test whether RtcB can splice strands composed entirely of DNA. To first gauge RtcB’s ability to guanylylate a DNA 3′-PO4 end, we assayed the transfer of 32P-GMP from 10 µM [α32P]GTP to a set of unlabeled DNA3′p oligonucleotides (2.5 µM) of differing length, but identical 3′-terminal nucleobase sequence (Fig. 3A). The reaction products were resolved by PAGE to reveal a descending ladder of 32P-labeled DNAppG species according to the size of the DNA strand (Fig. 3A). The yield of DNAppG did not vary significantly with DNA chain length from 23 nucleotides (75% of input DNAp radiolabeled with GMP) to 8 nucleotides (71% of input DNAp guanylylated) (Fig. 3A, Left), but declined progressively as the DNAp was shortened to 7 nucleotides (25%), 6 nucleotides (22%), and 5 nucleotides (8%) (Fig. 3A, Right).
Fig. 3.
Guanylylation of DNA 3′-PO4 ends by RtcB. (A) Reaction mixtures (10 µL) containing 50 mM Tris⋅HCl (pH 8.0), 2 mM MnCl2, 10 µM [α32P]GTP, and 5 µM RtcB were preincubated for 1 min at 37 °C, then supplemented with 25 pmol (2.5 µM) of unlabeled DNA3′p oligonucleotides of varying length as specified. After further incubation for 5 min at 37 °C, the reactions were quenched with 10 µL of 90% formamide, 50 mM EDTA, and the products were analyzed by urea-PAGE and visualized by autoradiography. The nucleobase sequences of the DNAp strands are shown (Lower). (B) Reaction mixture containing 50 mM Tris⋅HCl (pH 8.0), 2 mM MnCl2, 100 µM GTP, 4 µM RtcB, and 0.05 µM pD12p (shown with the 5′ 32P-label denoted by •) was incubated at 37 °C. Aliquots were withdrawn at the times specified and quenched immediately. The products were analyzed by urea-PAGE and visualized by autoradiography (Upper). The positions of the substrate and guanylylated product are indicated (Right). The kinetic profiles of the pD12p guanylylation reaction at three concentrations of input RtcB are plotted (Lower). Each datum in the graph is the average of three separate experiments ±SEM. Nonlinear regression curve fits of the data to a single exponential are shown.
Further analysis of DNAp guanylylation was performed by using a 5′ 32P-labeled, 3′-PO4 terminated DNA strand, pD12p (Fig. 3B). Under single-turnover conditions in the presence of 100 µM GTP and Mn2+, RtcB effected quantitative conversion of pD12p to pD12ppG (Fig. 3B, Upper). The rate of single-turnover DNAp guanylylation increased with RtcB concentration in the range of 0.5–4 µM (Fig. 3B, Lower), signifying that RtcB⋅DNAp association, rather than reaction chemistry, was rate limiting under these conditions. The observed rate constant for DNAp guanylylation by 4 µM RtcB was 5.0 ± 0.65 min−1. This value is similar to the observed rate constant of 7.1 min−1 for single-turnover guanylylation of the ribonucleoside-3′-PO4 terminus during ligation of the D17Rp substrate (Fig. 1B). The high efficiency with which RtcB guanylylates a DNAp end in vitro raises the prospect that RtcB family members could serve as DNA 3′ capping enzymes, conceptually analogous to RNA capping enzymes that add GMP to the 5′ ends of eukaryal mRNA. In such a scenario, the installation of a DNAppG cap could mark DNA for downstream transactions. At the very least, a DNAppG cap would protect the DNA strand from resection by 3′ exonucleases.
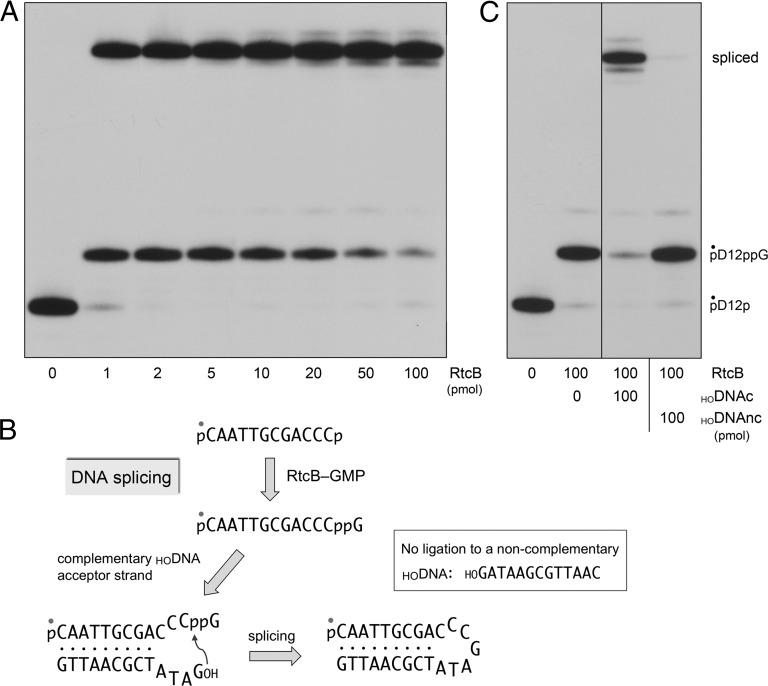
To assay DNA splicing, we reacted RtcB with a broken DNA stem-loop substrate that mimics the broken tRNA stem loops that RtcB can splice and repair in vivo (2). The DNA stem loop (shown in Fig. 4B) was formed by mixing the 5′ 32P-labeled pD12p strand with excess complementary unlabeled 13-mer 5′-OH DNA (HODNAc). In reactions containing 1 pmol (0.1 µM) of labeled DNA, 1 pmol (0.1 µM) of RtcB consumed nearly all of the substrate, generating a mixture of guanylylated intermediate (27%) and spliced product (70%) (Fig. 4A). Higher levels of input RtcB increased the yield of spliced DNA to 95% (Fig. 4A). An instructive finding was that RtcB was unable to ligate the activated pD12ppG intermediate to a noncomplementary 13-mer 5′-OH DNA (HODNAnc) (Fig. 4C). These experiments show that RtcB is indeed a DNA joining enzyme, aided by intermolecular complementarity to capture the 5′-OH DNA nucleophile for the final step of phosphodiester synthesis (Fig. 4B).
Fig. 4.
Splicing of DNA 3′-PO4 and 5′-OH ends by RtcB. (A) Reaction mixtures (10 µL) containing 50 mM Tris⋅HCl (pH 8.0), 2 mM MnCl2, 200 µM GTP, 1 pmol (0.1 µM) 5′ 32P-labeled pD12p substrate, 50 pmol (5 µM) of a complementary HODNA acceptor strand (HOGATATCGCAATTG), and RtcB as specified were incubated at 37 °C for 20 min. The products were analyzed by urea-PAGE and visualized by autoradiography. (B) Schematic diagram of the pathway of DNA splicing by RtcB. (C) Reaction mixtures (10 µL) containing 50 mM Tris⋅HCl (pH 8.0), 2 mM MnCl2, 200 µM GTP, 1 pmol (0.1 µM) 5′ 32P-labeled pD12p, 0 or 100 pmol (10 µM) RtcB, and 100 pmol (10 µM) of complementary (HODNAc) or noncomplementary (HODNAnc) DNA acceptors strand as indicated were incubated at 37 °C for 20 min. The products were analyzed by urea-PAGE and visualized by autoradiography. The positions of the pD12p substrate, pD12ppG intermediate, and spliced product are indicated (Right).
Intermediacy of DNA3′pp5′G in DNA Splicing.
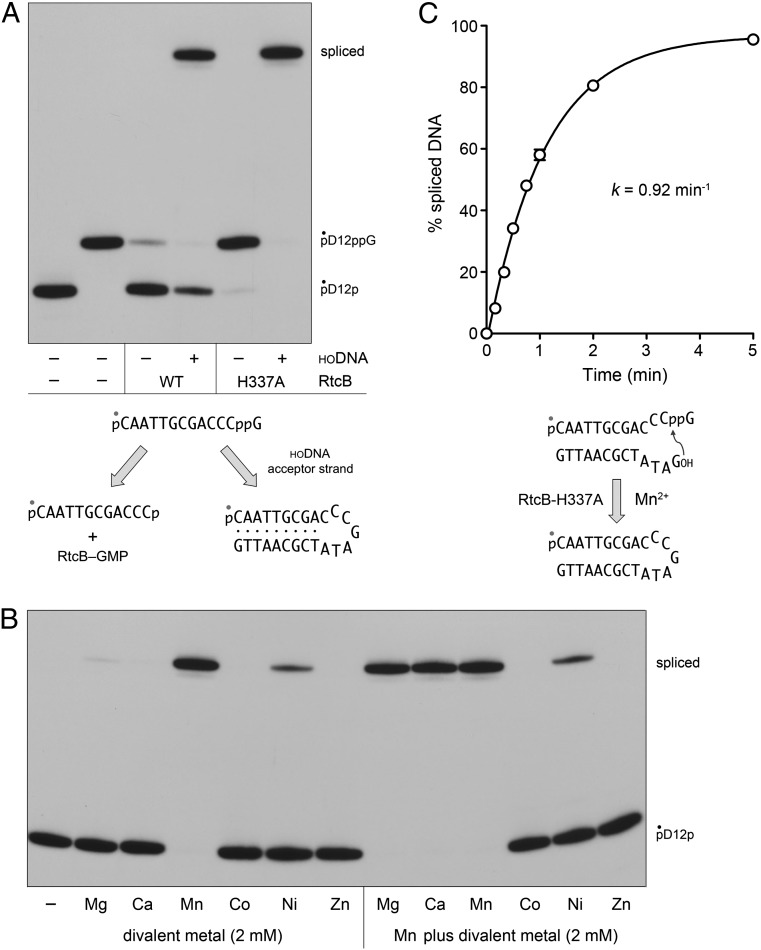
Whereas our analysis of single-turnover 3′-PO4 and 5′-OH splicing by RtcB argues strongly for a precursor–product relationship between the 3′-guanylylated species and the spliced species, the definitive test of intermediacy is provided by the experiments in Fig. 5A, in which we purified the 32P-labeled pD12ppG formed by reaction of RtcB with pD12p and GTP, and then reacted pD12ppG with RtcB and Mn2+ under single-turnover conditions (in the absence of GTP), with or without a complementary HODNA acceptor. Absent an acceptor strand, RtcB catalyzed near-quantitative deguanylylation of pDNAppG, generating pDNAp (97% yield). There are two potential mechanisms for the deguanylylation reaction: (i) microscopic reversal of the forward step of GMP transfer from the RtcB-(histdinyl337)–GMP intermediate to the 3′-PO4 of pD12p, whereby the His337 nucleophile attacks the guanylate phosphorus of pDNAppG to generate pD12p and RtcB–GMP; or (ii) direct hydrolysis of the 3′–5′ phosphoanhydride to form pD12p and GMP. The finding that the deguanylylation reaction was virtually abolished by mutation of His337 to alanine (Fig. 5A) weighs in favor of mechanism i. Consistent with mechanism i, we found that reaction of wild-type RtcB with gel-purified GMP-labeled DNAppG resulted in transfer of the 32P-guanylate to the enzyme to form an RtcB-guanylate adduct detectable by SDS/PAGE; no label transfer was detected to the H337A mutant (Fig. S2).
Fig. 5.
Reactions of RtcB with DNAppG and 5′-OH DNA. (A) Deguanylylation and phosphodiester synthesis. Reaction mixtures (10 µL) containing 50 mM Tris⋅HCl (pH 8.0,) 2 mM MnCl2, 0.05 µM 5′ 32P-labeled pD12ppG substrate, 1 µM wild-type (WT) RtcB or H337A mutant (where indicated by +), and 5 µM complementary HODNA acceptor strand HOGATATCGCAATTG (where indicated by +), were incubated at 37 °C for 20 min. The products were analyzed by urea-PAGE and visualized by autoradiography. The distinct deguanylylation and splicing reactions of RtcB with the guanylylated DNA are depicted (Lower). (B) Metal requirement and specificity in the isolated phosphodiester synthesis step. Reaction mixtures (10 µL) containing 50 mM Tris⋅HCl (pH 8.0), 0.05 µM 5′ 32P-labeled pD12ppG substrate, 5 µM HODNAc acceptor, 1 µM RtcB-H337A, and either no divalent metal (−), 2 mM of the indicated divalent cation (reaction series at Left) or a mixture of 2 mM MnCl2 plus 2 mM of the indicated divalent cation (reaction series at Right) were incubated at 37 °C for 20 min. The products were analyzed by urea-PAGE and visualized by autoradiography. (C) Kinetics of phosphodiester synthesis by RtcB-H337A. A reaction mixture containing 50 mM Tris⋅HCl (pH 8.0), 2 mM MnCl2, 0.05 µM 5′ 32P-labeled pD12ppG substrate, 5 µM HODNAc acceptor, and 1 µM RtcB-H337A was incubated at 37 °C. Aliquots were withdrawn at the times specified and quenched immediately. The kinetic profile of the phosphodiester synthesis reaction (Lower) is plotted in the graph, where each datum is the average of three separate experiments ±SEM. A nonlinear regression curve fit of the data to a single exponential is shown.
Provision of a HODNA acceptor attenuated the deguanylylation reaction (22% yield of pD12p) and resulted in the splicing of the pD12ppG and HODNA ends (76% yield) (Fig. 5A). Thus, the fate of pD12ppG reflects partitioning between forward phosphodiester synthesis and backward GMP transfer to RtcB, with the forward splicing step being favored under the conditions studied presently. The salient and instructive finding was that the RtcB H337A mutant, which is inert in joining 3′-PO4 and 5′-OH ends (6), catalyzed quantitative splicing of the pD12ppG and HODNA ends (Fig. 5A). Thus, the 3′-guanylylated species is a bona fide intermediate in 3′-PO4 to 5′-OH DNA splicing and there is no requirement for GTP or for His337 subsequent to the 3′-guanylylation step.
We exploited the H337A mutant and the isolated pD12ppG intermediate to gauge the requirements for the phosphodiester synthesis step, which was stringently dependent on a divalent cation. This requirement was satisfied by manganese, and to a feeble extent by nickel, but not by magnesium, calcium, cobalt, or zinc (Fig. 5B). A set of mixing experiments, in which reactions of H337A with pD12ppG and HODNA containing manganese were supplemented with an equivalent concentration of magnesium, calcium, cobalt, or zinc, was informative regarding metal specificity. Magnesium and calcium had no effect on the yield of spliced DNA sustained by manganese. By contrast, cobalt and zinc effaced all splicing activity in the presence of manganese (Fig. 5B). The extent of DNA splicing in the presence of nickel plus manganese was the same as that with nickel alone (Fig. 5B). These results for the isolated step of phosphodiester synthesis mirror the metal requirements and metal-inhibitory effects observed for the isolated reaction of RtcB with GTP to form the covalent RtcB–GMP intermediate (5). They support our initial hypothesis that RtcB binds its metal cofactor via “soft” enzymic ligands such as histidine nitrogens and a cysteine sulfur, such that “hard” metals like calcium and magnesium do not bind the active site, whereas soft metals such as zinc and cobalt do bind the active site (and out-compete manganese), but when so engaged are unable to sustain catalysis (1). This model is fortified by recent crystallographic evidence that RtcB binds two Mn2+ ions in coordination complexes populated by soft ligands (20, 21). A kinetic analysis of the isolated phosphodiester synthesis step catalyzed by the H337A mutant under conditions of enzyme excess is shown in Fig. 5C; the apparent rate constant of 0.92 ± 0.07 min−1 for sealing of a DNAppG end was similar to the value of 1.8 min−1 for attack of a DNA 5′-OH on a ribonucleotide-3′-ppG intermediate (Fig. 1B).
Conclusions and Implications
The present studies overturn the textbook version of enzymatic DNA end joining by instating a unique pathway for splicing of DNA 3′-PO4 (or cyclic-PO4) ends to DNA 5′-OH ends via a capped DNAppG intermediate. The implications of this mechanism for DNA break repair and DNA rearrangements are potentially significant, given the many biological settings in which DNA damage generates 3′-PO4 (or cyclic-PO4) and 5′-OH ends that are refractory to the action of conventional DNA ligases. Because RtcB homologs are present in the proteomes of scores of bacterial taxa, and bacteria have no evident need for protein-catalyzed tRNA splicing (the function ascribed to metazoan and archaeal RtcB homologs), there is no reason to limit our thinking about RtcB function in RNA versus DNA repair. Indeed, the fact that some bacteria encode multiple RtcB paralogs (e.g., Myxococcus xanthus encodes six distinct RtcB proteins) raises the prospect that RtcB-type ligases might acquire functional specialization for particular repair pathways or for the sealing of specific nucleic acid substrates (a scenario akin to the specialization of conventional DNA ligases in eukarya and bacteria).
Our studies here of DNA end joining by E. coli RtcB exploited an all-DNA stem-loop substrate that mimics the incised tRNA anticodon stem loop that RtcB can splice in vivo (2). Conventional DNA ligases that act during DNA replication and excision repair have specifically evolved to seal 3′-OH/5′-PO4 nicks in duplex DNA. When we interrogated the action of E. coli RtcB at a centrally placed 3′-PO4/5′-OH nick in a 36-bp duplex DNA, we found that the 3′-PO4 end was guanylylated, but there was no joining of the 5′ 32P-labeled DNAppG strand to the adjacent HODNA strand at the nick (Fig. S3). We surmise that one or both of the reactive—N3′ppG and 5′-HON—termini must be unpaired for E. coli RtcB to execute the final step of phosphodiester synthesis.
Further experiments gauged the effect of duplex secondary structure at the 3′-PO4 end on sealing efficiency. The HOD17Rp single strand was readily converted by RtcB to a circular product via intramolecular sealing (94% yield at 0.25 µM RtcB; Fig. S4, Center). Annealing of HOD17Rp to a complementary 36-mer DNA to form an 18-bp duplex with an 18-nucleotide 5′-OH single-stranded tail suppressed circularization (as expected), and allowed for inefficient joining of the duplex 3′-PO4 end to the 5′-OH of the tailed duplex to form a stem-loop product (2% and 15% yield at 1 and 4 µM RtcB, respectively; Fig. S4, Left). These findings indicate that E. coli RtcB is optimized to seal when the 3′-PO4 end is single stranded. RtcB was similarly feeble at joining HOD17Rp to a 5′-OH acceptor DNA in the context of a nicked duplex (1% and 4% yield at 1 and 4 µM RtcB, respectively; Fig. S4, Right).
If this is an immutable property of the DNA splicing reactions of all RtcB enzymes, then it suggests a potential role for RtcB in DNA rearrangements at double-strand breaks with single-stranded tails. Alternatively, certain RtcB homologs from taxa other than E. coli may be naturally adept at sealing 3′-PO4/5′-OH nicks in duplex DNA. Whereas the approximation of single-stranded ends can be achieved in model substrates by forming a duplex stem, it is conceivable that RtcB is assisted as an agent of nucleic acid rearrangements by partner proteins that bridge the ends (akin to the role of Ku in promoting ligation during nonhomologous end joining).
Finally, our findings highlight RtcB as a powerful and exquisitely specific reagent for tagging, isolating, amplifying, and/or sequencing DNAs and RNAs with 3′-PO4 or 2′,3′-cyclic-PO4 ends.
Methods
RtcB.
RtcB proteins were produced in E. coli and purified from soluble bacterial extracts as described (6). Protein concentrations were determined by the BioRad dye binding method with BSA as the standard.
Substrates.
The 3′ 32P-labeled D17Rp strand (Fig. 1A) was prepared as follows. A 10-mer deoxyribonucleotide HOATTCCGATAGOH was 5′ 32P-labeled by using [γ32P]ATP and T4 polynucleotide kinase-phosphatase (Pnkp), and then gel purified. The labeled pD10 strand was mixed with an 18-mer CATATCCGTGTCGCCCT(rC)OH strand (D17R) and a complementary 36-mer DNA strand TGTAGTCACTATCGGAATGAGGGCGACACGGATATG to form a nicked DNA duplex with a monoribonucleotide on the 3′-OH side of the nick and a 32P-labeled deoxynucleotide on the 5′-PO4 side of the nick. The nick was ligated by reaction with a 10-fold molar excess of Chlorella virus DNA ligase, 10 mM MgCl2, and 1 mM ATP for 1 h at 22 °C to yield an internally 32P-labeled D17RpD10 strand. The ligation product was heated for 10 min at 95 °C in the presence of a 10-fold molar excess of an unlabeled 36-mer DNA that competed with the radiolabeled ligation product for reannealing to the bridging template DNA strand. This mixture was digested with 50 µg RNase A for 10 min at 37 °C, thereby converting the D17RpD10 species to a 3′ 32P-labeled D17Rp strand, which was gel purified before use in strand joining assays. D17Rp was converted to D17R>p (Fig. S1A) by reaction with RNA 3′-terminal cyclase as described (22), then recovered by phenol:chloroform extraction and precipitation with ethanol. Quantitative cyclization of the 3′-PO4 was verified by treating the D17Rp and D17R>p strands with alkaline phosphatase followed by TLC analysis, which showed that all of the 32P-label in the D17Rp strand was liberated as 32Pi, whereas the label in the D17R>p strand was completely resistant to hydrolysis, as expected for a phosphodiester (22).
The 5′ 32P-labeled pD11Rp strand (Fig. 2) was prepared as follows. A 18-mer oligonucleotide CGTGTCGCCCT(rU)ATTCCC composed of 17 deoxynucleotides and a single internal ribouridine was 5′ 32P-labeled by using [γ32P]ATP and T4 Pnkp. The kinase was inactivated by heating the mixture at 95 °C for 3 min. The product was then digested with 50 µg RNase A for 10 min at 37 °C, thereby converting pCGTGTCGCCCT(rU)ATTCCC to pCGTGTCGCCCT(rU)p, which was gel purified before use in strand joining assays.
The 5′ 32P-labeled pD12p strand (Figs. 3 and 4) was prepared by enzymatic phosphorylation of a 12-mer HOCAATTGCGACCCp oligonucleotide by using [γ32P]ATP and the phosphatase-dead mutant T4 Pnkp-D167N. The radiolabeled pCAATTGCGACCCp strand was gel purified before use in DNA splicing and 3′-guanylylation assays. The 5′ 32P-labeled pD12ppG strand (Fig. 5) was prepared by incubating pD12p with a 20-fold molar excess of RtcB in the presence of 2 mM MnCl2 and 1 mM GTP for 20 min at 37 °C, followed by gel purification.
End Joining Assay.
Reaction mixtures containing 50 mM Tris⋅HCl (pH 8.0), 2 mM MnCl2, 100 μM GTP, and nucleic acid substrates and RtcB as specified were incubated at 37 °C. The reactions were quenched by adding an equal volume of 90% formamide, 50 mM EDTA. The products were resolved by electrophoresis at 55 W through a 40-cm 20% polyacrylamide gel containing 8 M urea in 45 mM Tris-borate, 1.2 mM EDTA. The 32P-labeled oligonucleotides were visualized by autoradiography and quantified by scanning the gel with a FujiFilm BAS-2500 imager.
Supplementary Material
Acknowledgments
This research was supported by National Institutes of Health Grant GM46330. S.S. is an American Cancer Society Research Professor.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 20354.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1314289110/-/DCSupplemental.
References
- 1.Tanaka N, Shuman S. RtcB is the RNA ligase component of an Escherichia coli RNA repair operon. J Biol Chem. 2011;286(10):7727–7731. doi: 10.1074/jbc.C111.219022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tanaka N, Meineke B, Shuman S. RtcB, a novel RNA ligase, can catalyze tRNA splicing and HAC1 mRNA splicing in vivo. J Biol Chem. 2011;286(35):30253–30257. doi: 10.1074/jbc.C111.274597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Englert M, Sheppard K, Aslanian A, Yates JR, 3rd, Söll D. Archaeal 3′-phosphate RNA splicing ligase characterization identifies the missing component in tRNA maturation. Proc Natl Acad Sci USA. 2011;108(4):1290–1295. doi: 10.1073/pnas.1018307108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Popow J, et al. HSPC117 is the essential subunit of a human tRNA splicing ligase complex. Science. 2011;331(6018):760–764. doi: 10.1126/science.1197847. [DOI] [PubMed] [Google Scholar]
- 5.Tanaka N, Chakravarty AK, Maughan B, Shuman S. Novel mechanism of RNA repair by RtcB via sequential 2′,3′-cyclic phosphodiesterase and 3′-Phosphate/5′-hydroxyl ligation reactions. J Biol Chem. 2011;286(50):43134–43143. doi: 10.1074/jbc.M111.302133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chakravarty AK, Subbotin R, Chait BT, Shuman S. RNA ligase RtcB splices 3′-phosphate and 5′-OH ends via covalent RtcB-(histidinyl)-GMP and polynucleotide-(3′)pp(5′)G intermediates. Proc Natl Acad Sci USA. 2012;109(16):6072–6077. doi: 10.1073/pnas.1201207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chakravarty AK, Shuman S. The sequential 2′,3′-cyclic phosphodiesterase and 3′-phosphate/5′-OH ligation steps of the RtcB RNA splicing pathway are GTP-dependent. Nucleic Acids Res. 2012;40(17):8558–8567. doi: 10.1093/nar/gks558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eastberg JH, Pelletier J, Stoddard BL. Recognition of DNA substrates by T4 bacteriophage polynucleotide kinase. Nucleic Acids Res. 2004;32(2):653–660. doi: 10.1093/nar/gkh212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garces F, Pearl LH, Oliver AW. The structural basis for substrate recognition by mammalian polynucleotide kinase 3′ phosphatase. Mol Cell. 2011;44(3):385–396. doi: 10.1016/j.molcel.2011.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coquelle N, Havali-Shahriari Z, Bernstein N, Green R, Glover JNM. Structural basis for the phosphatase activity of PNKP on single- and double-stranded DNA substrates. Proc Natl Acad Sci USA. 2011;108:21022–21027. doi: 10.1073/pnas.1112036108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang LK, Das U, Smith P, Shuman S. Structure and mechanism of the polynucleotide kinase component of the bacterial Pnkp-Hen1 RNA repair system. RNA. 2012;18(12):2277–2286. doi: 10.1261/rna.036061.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nair PA, Smith P, Shuman S. Structure of bacterial LigD 3′-phosphoesterase unveils a DNA repair superfamily. Proc Natl Acad Sci USA. 2010;107(29):12822–12827. doi: 10.1073/pnas.1005830107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith P, Nair PA, Das U, Zhu H, Shuman S. Structures and activities of archaeal members of the LigD 3′-phosphoesterase DNA repair enzyme superfamily. Nucleic Acids Res. 2011;39(8):3310–3320. doi: 10.1093/nar/gkq1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das U, Shuman S. Mechanism of RNA 2′,3′-cyclic phosphate end healing by T4 polynucleotide kinase-phosphatase. Nucleic Acids Res. 2013;41(1):355–365. doi: 10.1093/nar/gks977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sekiguchi J, Shuman S. Site-specific ribonuclease activity of eukaryotic DNA topoisomerase I. Mol Cell. 1997;1(1):89–97. doi: 10.1016/s1097-2765(00)80010-6. [DOI] [PubMed] [Google Scholar]
- 16.Reijns MAM, et al. Enzymatic removal of ribonucleotides from DNA is essential for mammalian genome integrity and development. Cell. 2012;149(5):1008–1022. doi: 10.1016/j.cell.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim N, et al. Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I. Science. 2011;332(6037):1561–1564. doi: 10.1126/science.1205016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sparks JL, et al. RNase H2-initiated ribonucleotide excision repair. Mol Cell. 2012;47(6):980–986. doi: 10.1016/j.molcel.2012.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williams JS, et al. Topoisomerase 1-mediated removal of ribonucleotides from nascent leading-strand DNA. Mol Cell. 2013;49(5):1010–1015. doi: 10.1016/j.molcel.2012.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Desai KK, Bingman CA, Phillips GN, Jr, Raines RT. Structures of the noncanonical RNA ligase RtcB reveal the mechanism of histidine guanylylation. Biochemistry. 2013;52(15):2518–2525. doi: 10.1021/bi4002375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Englert M, et al. Structural and mechanistic insights into guanylylation of RNA-splicing ligase RtcB joining RNA between 3′-terminal phosphate and 5′-OH. Proc Natl Acad Sci USA. 2012;109(38):15235–15240. doi: 10.1073/pnas.1213795109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Das U, Shuman S. 2′-Phosphate cyclase activity of RtcA: A potential rationale for the operon organization of RtcA with an RNA repair ligase RtcB in Escherichia coli and other bacterial taxa. RNA. 2013;19(10):1355–1362. doi: 10.1261/rna.039917.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.