Abstract
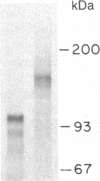
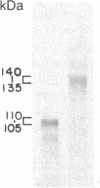
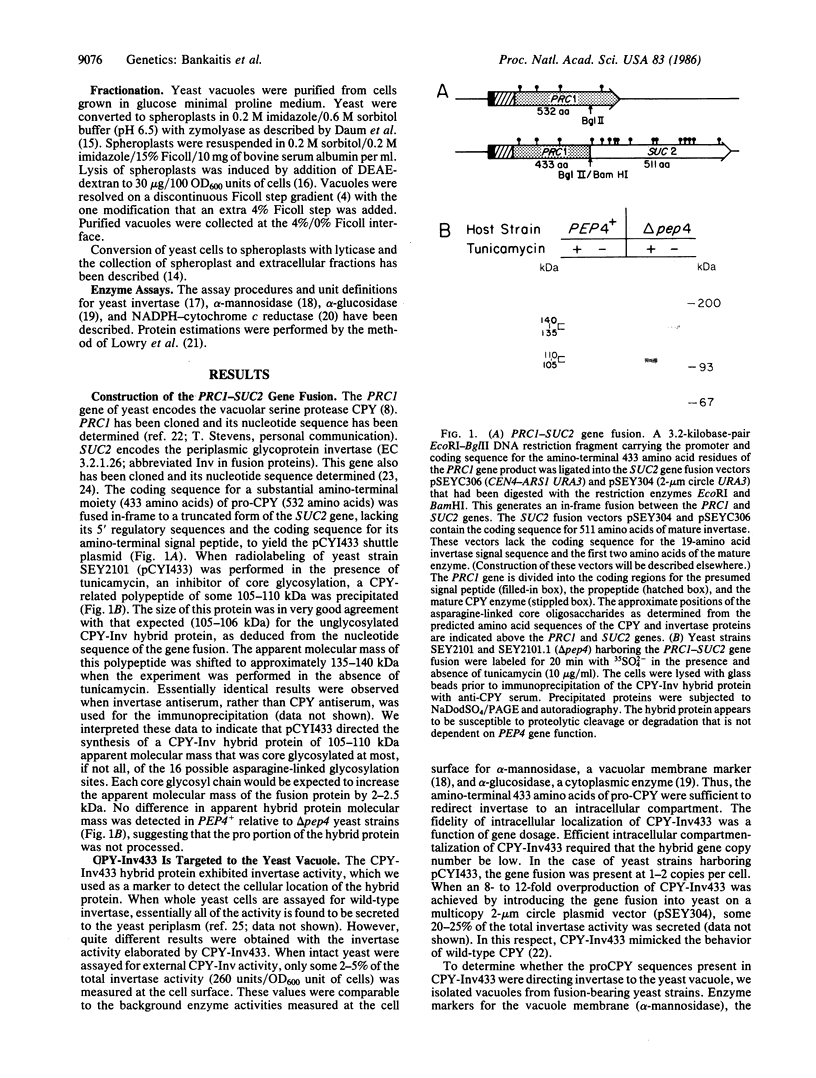
We have constructed a PRC1-SUC2 gene fusion that directs the synthesis in Saccharomyces cerevisiae of a hybrid polypeptide consisting of a 433-residue amino-terminal domain derived from the yeast vacuolar protease carboxypeptidase Y (CPY; EC 3.4.16.1) and a 511-residue carboxyl-terminal domain derived from the secreted yeast enzyme invertase (EC 3.2.1.26). Fractionation data indicated that this amount of CPY primary sequence is sufficient to quantitatively divert invertase to the yeast vacuole. The phenotypic consequence of localizing active invertase to the vacuole has enabled us to select for mutants that "mislocalize" the hybrid protein to the cell surface. The corresponding mutations that lead to this effect are all trans-acting and recessive, and they define at least eight complementation groups. These vacuolar protein targeting (vpt) mutants also exhibit hybrid protein independent defects in wild-type CPY delivery to the yeast vacuole. Precursor forms of CPY accumulate in the mutants and are secreted into the yeast periplasm and extracellular medium. The vpt mutants should provide useful information pertaining to the mechanisms by which yeast cells regulate vacuolar protein traffic.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Carlson M., Botstein D. Two differentially regulated mRNAs with different 5' ends encode secreted with intracellular forms of yeast invertase. Cell. 1982 Jan;28(1):145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- Daum G., Böhni P. C., Schatz G. Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J Biol Chem. 1982 Nov 10;257(21):13028–13033. [PubMed] [Google Scholar]
- Douglas M. G., Geller B. L., Emr S. D. Intracellular targeting and import of an F1-ATPase beta-subunit-beta-galactosidase hybrid protein into yeast mitochondria. Proc Natl Acad Sci U S A. 1984 Jul;81(13):3983–3987. doi: 10.1073/pnas.81.13.3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dürr M., Boller T., Wiemken A. Polybase induced lysis of yeast spheroplasts. A new gentle method for preparation of vacuoles. Arch Microbiol. 1975 Nov 7;105(3):319–327. doi: 10.1007/BF00447152. [DOI] [PubMed] [Google Scholar]
- Emr S. D., Schekman R., Flessel M. C., Thorner J. An MF alpha 1-SUC2 (alpha-factor-invertase) gene fusion for study of protein localization and gene expression in yeast. Proc Natl Acad Sci U S A. 1983 Dec;80(23):7080–7084. doi: 10.1073/pnas.80.23.7080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein A., Lampen J. O. Beta-D-fructofuranoside fructohydrolase from yeast. Methods Enzymol. 1975;42:504–511. doi: 10.1016/0076-6879(75)42159-0. [DOI] [PubMed] [Google Scholar]
- HALVORSON H., ELLIAS L. The purification and properties of an alpha-glucosidase of Saccharomyces italicus Y1225. Biochim Biophys Acta. 1958 Oct;30(1):28–40. doi: 10.1016/0006-3002(58)90237-3. [DOI] [PubMed] [Google Scholar]
- Hall M. N., Hereford L., Herskowitz I. Targeting of E. coli beta-galactosidase to the nucleus in yeast. Cell. 1984 Apr;36(4):1057–1065. doi: 10.1016/0092-8674(84)90055-2. [DOI] [PubMed] [Google Scholar]
- Hasilik A., Tanner W. Biosynthesis of the vacuolar yeast glycoprotein carboxypeptidase Y. Conversion of precursor into the enzyme. Eur J Biochem. 1978 Apr 17;85(2):599–608. doi: 10.1111/j.1432-1033.1978.tb12275.x. [DOI] [PubMed] [Google Scholar]
- Hasilik A., Tanner W. Carbohydrate moiety of carboxypeptidase Y and perturbation of its biosynthesis. Eur J Biochem. 1978 Nov 15;91(2):567–575. doi: 10.1111/j.1432-1033.1978.tb12710.x. [DOI] [PubMed] [Google Scholar]
- Hemmings B. A., Zubenko G. S., Hasilik A., Jones E. W. Mutant defective in processing of an enzyme located in the lysosome-like vacuole of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1981 Jan;78(1):435–439. doi: 10.1073/pnas.78.1.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin J. V., Adams A. E. Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces. J Cell Biol. 1984 Mar;98(3):922–933. doi: 10.1083/jcb.98.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota S., Yoshida Y., Kumaoka H., Furumichi A. Studies on the microsomal electron-transport system of anaerobically grown yeast. V. Purification and characterization of NADPH-cytochrome c reductase. J Biochem. 1977 Jan;81(1):197–205. doi: 10.1093/oxfordjournals.jbchem.a131436. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Novick P., Ferro S., Schekman R. Order of events in the yeast secretory pathway. Cell. 1981 Aug;25(2):461–469. doi: 10.1016/0092-8674(81)90064-7. [DOI] [PubMed] [Google Scholar]
- Novick P., Field C., Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980 Aug;21(1):205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- Novick P., Schekman R. Export of major cell surface proteins is blocked in yeast secretory mutants. J Cell Biol. 1983 Feb;96(2):541–547. doi: 10.1083/jcb.96.2.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opheim D. J. alpha-D-Mannosidase of Saccharomyces cerevisiae. Characterization and modulation of activity. Biochim Biophys Acta. 1978 May 11;524(1):121–130. doi: 10.1016/0005-2744(78)90110-9. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Schauer I., Emr S., Gross C., Schekman R. Invertase signal and mature sequence substitutions that delay intercompartmental transport of active enzyme. J Cell Biol. 1985 May;100(5):1664–1675. doi: 10.1083/jcb.100.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaiger H., Hasilik A., von Figura K., Wiemken A., Tanner W. Carbohydrate-free carboxypeptidase Y is transferred into the lysosome-like yeast vacuole. Biochem Biophys Res Commun. 1982 Feb 11;104(3):950–956. doi: 10.1016/0006-291x(82)91341-9. [DOI] [PubMed] [Google Scholar]
- Silhavy T. J., Beckwith J. Isolation and characterization of mutants of Escherichia coli K12 affected in protein localization. Methods Enzymol. 1983;97:11–40. doi: 10.1016/0076-6879(83)97115-x. [DOI] [PubMed] [Google Scholar]
- Stevens T. H., Rothman J. H., Payne G. S., Schekman R. Gene dosage-dependent secretion of yeast vacuolar carboxypeptidase Y. J Cell Biol. 1986 May;102(5):1551–1557. doi: 10.1083/jcb.102.5.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens T., Esmon B., Schekman R. Early stages in the yeast secretory pathway are required for transport of carboxypeptidase Y to the vacuole. Cell. 1982 Sep;30(2):439–448. doi: 10.1016/0092-8674(82)90241-0. [DOI] [PubMed] [Google Scholar]
- Taussig R., Carlson M. Nucleotide sequence of the yeast SUC2 gene for invertase. Nucleic Acids Res. 1983 Mar 25;11(6):1943–1954. doi: 10.1093/nar/11.6.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]