Abstract
PRUNE2 plays an important role in regulating tumor cell differentiation, proliferation, and invasiveness in neuroblastoma. Our previous study revealed that PRUNE2/OBSCN two-gene relative expression classifer accurately differentiated leiomyosarcoma from gastrointestinal stromal tumor. However, the association between PRUNE2 expression and prognosis in leiomyosarcoma is poorly understood. In this study, we evaluated the prognostic role of PRUNE2 in leiomyosarcoma. PRUNE2 expression was detected using immunohistochemistry in 30 formalin-fixed, paraffin-embedded leiomyosarcoma tissues from MD Anderson Cancer Center, and high expression was detected in 36.7% (11/30) of the samples. To validate these results, immunohistochemistry was performed on another cohort of 45 formalin-fixed, paraffin-embedded leiomyosarcoma tissues from Tianjin Medical University Cancer Institute & Hospital, and high PRUNE2 protein expression was detected in 37.8% (17/45) of the samples. Moreover, elevated PRUNE2 expression was significantly associated with tumor size (P = 0.03) and hemorrhage/cyst (P = 0.014), and was an independent favorable prognostic factor for overall survival in leiomyosarcoma patients from Tianjin Medical University Cancer Institute & Hospital (P < 0.05). These data suggest that increased PRUNE2 protein expression may serve as a favorable prognostic marker in human leiomyosarcoma.
Keywords: Leiomyosarcoma, PRUNE2, survival, prognosis
Leiomyosarcoma is a rare malignant soft tissue tumor, accoun-ting for approximately 10% of soft tissue sarcomas[1]. These tumors occur primarily in the uterus and gastrointestinal tract, and have phenotypic features of smooth muscle differentiation[2],[3]. Leiomyosarcoma is difficult to treat, and the prognosis for the disease is poor. PRUNE2 (prune homolog 2, Drosophila) is a susceptibility gene for Alzheimer disease and an important regulator of Rho signaling[4],[5]. High PRUNE2 expression has been reported in the nervous system and the brain of humans, as well as in the spinal cord of mice[5],[6]. We previously showed that PRUNE2/OBSCN two-gene relative expression classifer could be used as a biomarker to accurately distinguish leiomyosarcoma from gastrointestinal stromal tumors[7]. Recently, Machida et al.[6] reported that PRUNE2 regulates differentiation, proliferation, and invasion of neuroblastoma tumor cells and that increased expression of this protein is associated with favorable prognosis in human neuroblastomas. However, our previous data showed that PRUNE2 mRNA expression was not significantly associated with survival time in leiomyosarcoma patients[8]. Thus, the association between PRUNE2 expression and prognosis is poorly understood in leiomyosarcoma patients. To determine the prognostic role of PRUNE2 in leiomyosarcoma, we investigated the association between PRUNE2 protein expression and survival.
Materials and Methods
Patients and clinical information
We obtained formalin-fixed, paraffin-embedded tissue sections from 30 leiomyosarcoma patients at the University of Texas MD Anderson Cancer Center (MDACC; Houston, Texas, USA). These included samples from the retroperitoneum (n = 12), gastrointestinal tract (n = 9), uterus (n = 6), and skin (n = 3).
To validate the results obtained with the MDACC samples, another cohort of 45 formalin-fixed, paraffin-embedded tissues from patients with leiomyosarcoma were retrieved from Tianjin Medical University Cancer Institute & Hospital (TMUCIH) for further study. The patients' clinicopathologic characteristics, including sex, age, clinical stage, degree of cell differentiation, tumor location, tumor size, the presence or absence of hemorrhage/cyst, and histopathologic subtype, were also collected. The tissues had been acquired from 18 male and 27 female patients with a mean age of 55.8 years. Histopathologic subtypes were assigned based on the 2002 World Health Organization classification of tumors of soft tissue and bone[9],[10].
Overall survival time was defined from the date of diagnosis or the first treatment to the date of death or last follow-up. Overall survival ranged from 1 to 74 months, and the median survival was 19 months. The study protocol was approved by the institutional review boards of both hospitals, and consent was obtained from each patient.
Immunohistochemical (IHC) analysis
IHC staining was performed on all tissue samples using conventional methods. Briefly, tissue sections (4 µm) were dewaxed with xylene, rehydrated with ethanol, soaked in citrate buffer solution (10 mmol/L, pH=6), and boiled in a pressure cooker for 2.5 min to repair the antigen. Endogenous peroxidase activity was blocked with 3% H2O2 for 20 min. The sections were then blocked for 20 min with normal serum (Vector Laboratories, Burlingame, CA) and incubated overnight at 4°C with rabbit polyclonal anti-PRUNE2 antibody (Abcam Company, Abcam, Cambridge, UK; diluted 1:100). The antibody was substituted with PBS as a negative control. Secondary antibody was applied for 60 min (one drop of biotinylated anti-rabbit with three drops of normal rabbit serum). DAB (DAKO Corporation, Carpinteria, CA) staining was applied for 5–10 min and monitored by microscopy. Sections were counterstained for 30 s with hematoxylin and dehydrated with ethanol and xylene. Finally, all sections were mounted on coverslips.
Two pathologists blinded to clinical information evaluated and scored PRUNE2 staining based on staining extent and intensity[6]. Ten random high-power fields, each containing approximately 100 cells, were observed under the microscope (40×). First, staining extent was scored according to the proportion of positive tumor cells: 0% (score 0), ≤10% (score 1), 11% to 25% (score 2), 26% to 50% (score 3), 51% to 75% (score 4), and > 75% (score 5). Second, staining intensity was scored based on the color observed: no color (score 0), yellow (score 1), tan (score 2), and brown (score 3). Final scores were calculated by adding the extent and intensity scores, and the results were used to divide patients into two groups: low expression group, for final scores of 0–4, and high expression group, for final scores > 4.
Statistical analysis
SPSS statistical software version 16.0 for Windows was used for data analysis. Comparisons of frequencies were made with the chi-square test. Kaplan-Meier survival analysis (log-rank test) and multivariate Cox regression analysis were used to examine the relationship between overall survival and PRUNE2 protein expression. Two-tailed P values less than 0.05 were considered statistically significant.
Results
The protein expression and prognostic role of PRUNE2 in leiomyosarcoma samples from MDACC
In the 30 leiomyosarcoma tissues from MDACC, PRUNE2 protein staining was predominantly in the cytoplasm of cells (Figure 1). Low expression was discovered in 63.3% (19/30) of the samples (Figure 1A), while high expression was observed in 36.7% (11/30) (Figure 1B). To investigate the prognostic role of PRUNE2, we analyzed the association between PRUNE2 protein expression and overall survival. Univariate analysis showed that PRUNE2 protein expression had no significant association with overall survival (χ2 = 0.246, P = 0.62) (Figure 2A).
Figure 1. PRUNE2 protein expression in leiomyosarcoma tissues from MD Anderson Cancer Center under a representative high-power field (40×).
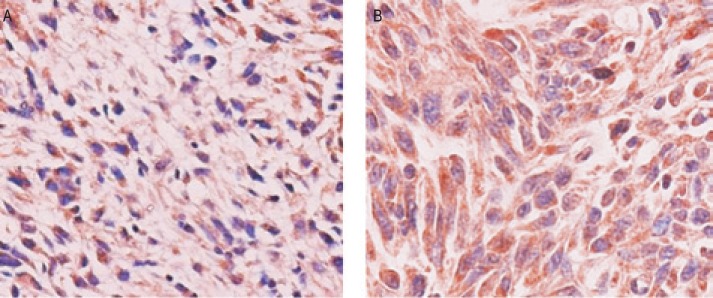
A, low expression of PRUNE2 protein; B, high expression of PRUNE2 protein.
Figure 2. The association between PRUNE2 protein expression and overall survival in patients with leiomyosarcoma.
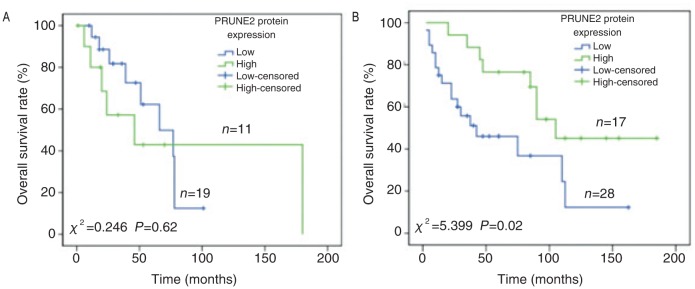
A, PRUNE2 protein expression is not significantly associated with overall survival in patients from MD Anderson Cancer Center; B, PRUNE2 protein expression is significantly associated with overall survival in patients from Tianjin Medical University Cancer Institute & Hospital.
The protein expression and prognostic role of PRUNE2 in leiomyosarcoma samples from TMUCIH
To validate these results and further evaluate the prognostic role of PRUNE2 in leiomyosarcoma, we used a larger cohort of leiomyosarcoma tissue samples from TMUCIH. In this set of 45 samples, PRUNE2 was also predominantly detected in the cytoplasm; and 62.2% (28/45) had low expression, while 37.8% (17/45) had high expression. We also analyzed the association between PRUNE2 protein expression and clinicopathologic factors, and found that PRUNE2 protein expression was significantly associated with tumor size (P = 0.03) and the absence of hemorrhage/cyst (P = 0.014) (Table 1). In addition, univariate analysis showed that there was a significant association between PRUNE2 protein expression and overall survival (χ2 = 5.399, P = 0.02) (Figure 2B), with longer survival noted among patients with high levels of the protein. Furthermore, multivariate analysis revealed that high PRUNE2 protein expression was an independent, favorable prognostic factor for overall survival (HR = 0.373, P = 0.025).
Table 1. Association between PRUNE2 protein expression and clinicopathologic factors of patients from Tianjin Medical University Cancer Institute & Hospital leiomyosarcoma samples.
| Characteristic | PRUNE2 expression |
χ2 | P | |
| Low | High | |||
| Sex | 0.252 | 0.616 | ||
| Male | 12 (26.7%) | 6 (13.3%) | ||
| Female | 16 (35.6%) | 11 (24.4%) | ||
| Age (years) | 0.733 | 0.392 | ||
| < 50 | 10 (22.2%) | 4 (8.9%) | ||
| ≥50 | 18 (40.0%) | 13 (28.9%) | ||
| Clinical stage | 0.180 | 0.672 | ||
| I-II | 13 (28.9%) | 9 (20.0%) | ||
| III-IV | 15 (33.3%) | 8 (17.8%) | ||
| Degree of differentiation | 0.044 | 0.978 | ||
| Poor | 5 (11.1%) | 3 (6.7%) | ||
| Moderate | 9 (20.0%) | 5 (11.1%) | ||
| Well | 14 (31.1%) | 9 (20.0%) | ||
| Tumor location | 2.567 | 0.277 | ||
| Retroperitoneal | 8 (17.8%) | 4 (8.9%) | ||
| Abdominopelvic | 10 (22.2%) | 10 (22.2%) | ||
| Visceral | 10 (22.2%) | 3 (6.7%) | ||
| Tumor size | 4.710 | 0.030 | ||
| < 10 cm | 14 (31.1%) | 14 (31.1%) | ||
| ≥10 cm | 14 (31.1%) | 3 (6.7%) | ||
| Hemorrhage/cyst | 6.035 | 0.014 | ||
| No | 17 (37.8%) | 16 (35.6%) | ||
| Yes | 11 (24.4%) | 1 (2.2%) | ||
| Histopathologic subtype | 0.093 | 0.955 | ||
| Classic | 19 (42.2%) | 11 (24.4%) | ||
| Epithelioid | 4 (8.9%) | 3 (6.7%) | ||
| Pleomorphic | 5 (11.1%) | 3 (6.7%) | ||
Discussion
The most important contribution of this study is that we found a significant association between PRUNE2 protein expression and overall survival, and that PRUNE2 protein expression was an independent prognostic factor for leiomyosarcoma. More specifically, leiomyosarcoma patients with higher PRUNE2 protein expression exhibited a better survival trend, suggesting that increased levels of the protein might be a favorable prognostic factor for leiomyosarcoma. Similarly, in neuroblastoma and prostate cancer, PRUNE2 protein is highly expressed and also plays a prognostic role[6],[11]. On the other hand, we found no statistically significant association between PRUNE2 mRNA expression and survival time in tissue samples from patients at MDACC in our previous study[8]. Here, Kaplan-Meier survival analysis revealed no significant relationship between overall survival time and PRUNE2 protein expression in samples from MDACC, which is consistent with our previous result[8]. Nevertheless, in the larger cohort of samples from TMUCIH, PRUNE2 protein expression showed a significant association with overall survival and was an independent prognostic factor. Thus, patients with higher PRUNE2 expression survived longer, indicating better prognosis. It is unclear why the results from MDACC and TMUCIH samples were different. Possible causes are the differences in race among the patients from each center and in the number of cases.
We also found that PRUNE2 protein expression was significantly associated with tumor size and hemorrhage/cyst. Specifically, PRUNE2 protein expression was higher in the smaller tumor size group (<10 cm). Because PRUNE2 plays an important role in regulating cell differentiation and apoptosis, this result suggests that patients with larger tumors may have worse survival.
Leiomyosarcoma is a rare soft tissue tumor that can occur anywhere in the human body. Several factors have been reported to predict poor outcomes in patients with leiomyosarcoma, including advanced age, vascular invasion, DNA aneuploidy, and c-Myc expression[2],[12],[13]. In particular, c-Myc expression was reported to be a marker for poor prognosis in leiomyosarcoma[13]. In this study, we have identified that PRUNE2 protein expression level can serve as a prognostic factor for leiomyosarcoma.
In conclusion, we report the association between PRUNE2 protein expression and prognosis in human leiomyosarcoma. Survival time was longer in TMUCIH leiomyosarcoma patients with higher expression of PRUNE2 protein. This suggests that PRUNE2 may be involved in the process of leiomyosarcoma development and can be regarded as a biomarker for favorable prognosis. Further study is necessary to better understand the role that racial factors play for the relationship between PRUNE2 protein expression and prognosis in patients with leiomyosarcoma, and discover new therapeutic strategies against aggressive leiomyosarcoma.
Acknowledgments
This work was partly supported by grants from the National Nature Science Foundation of China (No. 81372872 to J. Yang and No. 81320108022 to K. Chen), the University Cancer Foundation via the Sister Institution Network Fund (SINF) at the Tianjin Medical University Cancer Institute & Hospital (TMUCIH), Fudan University Shanghai Cancer Center (FUSCC) and University of Texas MD Anderson Cancer Center (UTMDACC), Changjiang Scholars and Innovative Research Team in University (PCSIRT) in China (No. IRT1076), and National Key Scientific and Technological Project (No. 2011ZX09037-001-04) (K. Chen).
References
- 1.Radkowski CA, Dodd LG, Johnson JL, et al. Leiomyosarcoma of the somatic soft tissues. J Surg Orthop Adv. 2012;21:96–101. doi: 10.3113/jsoa.2012.0096. [DOI] [PubMed] [Google Scholar]
- 2.Svarvar C, Bohling T, Berlin O, et al. Clinical course of nonvisceral soft tissue leiomyosarcoma in 225 patients from the scandinavian sarcoma group. Cancer. 2007;109:282–291. doi: 10.1002/cncr.22395. [DOI] [PubMed] [Google Scholar]
- 3.Mankin HJ, Casas-Ganem J, Kim JI, et al. Leiomyosarcoma of somatic soft tissues. Clin Orthop Relat Res. 2004;421:225–231. doi: 10.1097/01.blo.0000119250.08614.82. [DOI] [PubMed] [Google Scholar]
- 4.Potkin SG, Guffanti G, Lakatos A, et al. Hippocampal atrophy as a quantitative trait in a genome-wide association study identifying novel susceptibility genes for Alzheimer's disease. PloS One. 2009;4:e6501. doi: 10.1371/journal.pone.0006501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soh UJ, Low BC. BNIP2 extra long inhibits RhoA and cellular transformation by Lbc RhoGEF via its BCH domain. J Cell Sci. 2008;121:1739–1749. doi: 10.1242/jcs.021774. [DOI] [PubMed] [Google Scholar]
- 6.Machida T, Fujita T, Ooo ML, et al. Increased expression of proapoptotic BMCC1, a novel gene with the BNIP2 and Cdc42GAP homology (BCH) domain, is associated with favorable prognosis in human neuroblastomas. Oncogene. 2006;25:1931–1942. doi: 10.1038/sj.onc.1209225. [DOI] [PubMed] [Google Scholar]
- 7.Price ND, Trent J, El-Naggar AK, et al. Highly accurate two-gene classifier for differentiating gastrointestinal stromal tumors and leiomyosarcomas. Proc Natl Acad Sci USA. 2007;104:3414–3419. doi: 10.1073/pnas.0611373104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang JL, Cogdell D, Eddy J, et al. Expression of PRUNE2 mRNA and its positive correlation with non-coding RNA PCA3 in leiomyosarcoma. Zhonghua Zhong Liu Za Zhi. 2012;34:497–500. doi: 10.3760/cma.j.issn.0253-3766.2012.07.005. [in Chinese] [DOI] [PubMed] [Google Scholar]
- 9.Nicolas MM, Tamboli P, Gomez JA, et al. Pleomorphic and dedifferentiated leiomyosarcoma: clinicopathologic and immunohistochemical study of 41 cases. Hum Pathol. 2010;41:663–671. doi: 10.1016/j.humpath.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 10.Murphey MD. World Health Organization classification of bone and soft tissue tumors: modifications and implications for radiologists. Semin Musculoskelet Radiol. 2007;11:201–214. doi: 10.1055/s-2008-1038310. [DOI] [PubMed] [Google Scholar]
- 11.Clarke RA, Zhao Z, Guo AY, et al. New genomic structure for prostate cancer specific gene PCA3 within BMCC1: implications for prostate cancer detection and progression. PloS One. 2009;4:e4995. doi: 10.1371/journal.pone.0004995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gustafson P, Willen H, Baldetorp B, et al. Soft tissue leiomyo-sarcoma. A population-based epidemiologic and prognostic study of 48 patients, including cellular DNA content. Cancer. 1992;70:114–119. doi: 10.1002/1097-0142(19920701)70:1<114::aid-cncr2820700119>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 13.Tsiatis AC, Herceg ME, Keedy VL, et al. Prognostic significance of c-Myc expression in soft tissue leiomyosarcoma. Mod Pathol. 2009;22:1432–1438. doi: 10.1038/modpathol.2009.113. [DOI] [PubMed] [Google Scholar]


