Figure 3.
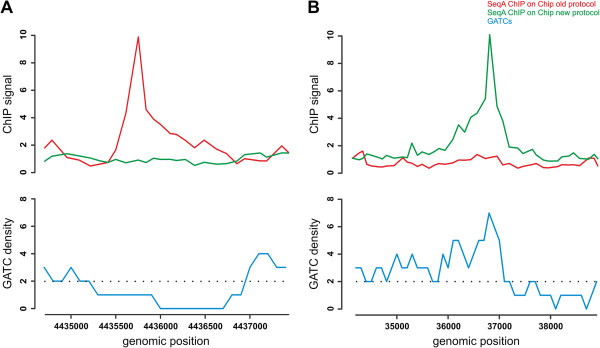
False positive enrichment peaks resemble true binding sites in ChIP-Chip experiments. Our modified ChIP-Chip protocol (green) resulted in enrichment data corresponding to the density of GATC binding sites (blue; moving window of 500 bp; step size 100 bp) as expected for SeqA binding with low signals in low GATC regions (A) and high signals in GATC dense chromosomal regions (B)[1]. The standard ChIP-Chip protocol (red) resulted in erroneous enrichment peaks in regions with low GATC density (A) and low signals in GATC rich regions (B). The shape and signal level of the false positive signal example in the ytfI gene region (red in A) and the true positive in the caiC gene region (green in B) are similar.
