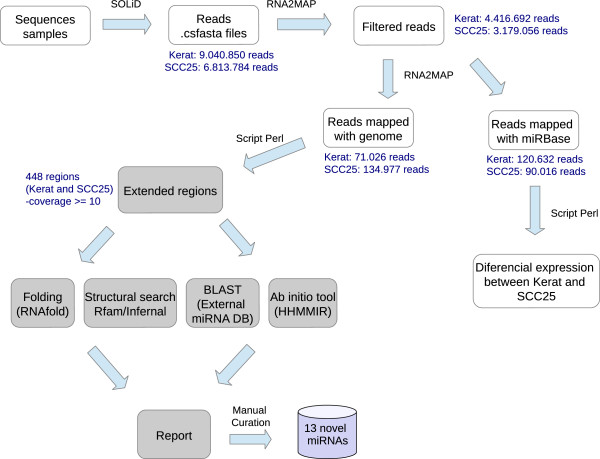
Figure 3.
Workflow for the analysis of small RNA transcriptome focusing on miRNA identification and discovery. Each library was converted to FASTA format, subjected to RNA2MAP tool and reads that did not match the human genome were discarded. After the filtering protocol, that matched miRBase were submitted to differential expression analysis and those that did not match were mapped to the genome. At each genomic locus the read was extended by100 nt up and downstream. Resulting sequences were subjected to the following pipeline: secondary structure prediction using RNAfold and Infernal against RFAM, blast against a local non-coding RNA database and, ab intio characterization using HHMMiR and RNAFold (HHMMiR in fact is performed using the data produced by RNAfold).