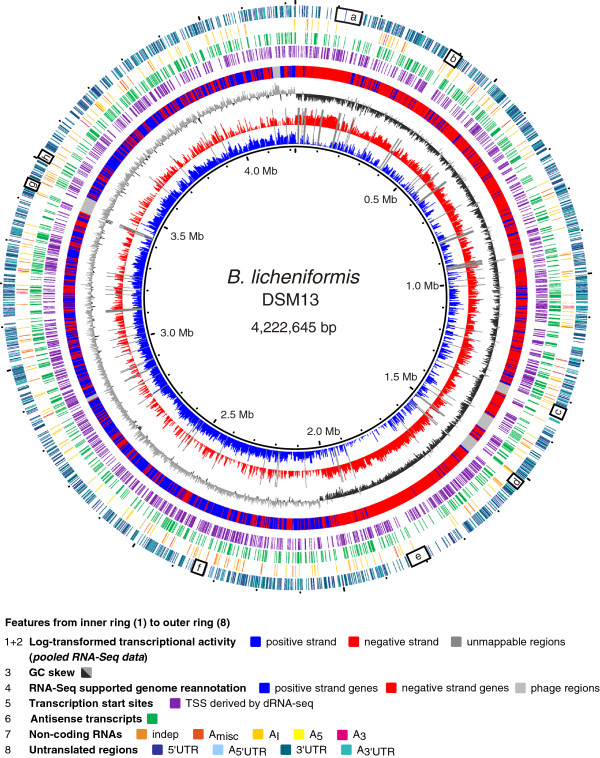
Figure 10.
Circular plot of transcriptional activity and identified RNA features. Combined depiction of reannotated genes and transcriptional activity of B. licheniformis. Unmappable regions, GC skew, transcription start sites, non-coding RNAs, untranslated regions and antisense transcripts are also shown. 5’UTRs and 3’UTRs are evenly distributed over the whole chromosome of B. licheniformis, except for regions a – h: these regions contain long operon structures (a: ribosomal superoperon, b: lch operon, e: fla/che operon, f: trp operon, h: eps operon) or prophage regions with low transcriptional activity (c, d and g). The classification scheme corresponds to Figure 4. (indep) ncRNA, not antisense to any mRNA; (Amisc) ncRNA partially antisense to an mRNA transcript or antisense to more than one gene; (Ai) ncRNA completely antisense to a protein-coding gene; (A5) ncRNA exclusively antisense to the 5'UTR of an mRNA; (A3) ncRNA exclusively antisense to the 5'UTR of an mRNA; (5'UTR) 5'untranslated region of an mRNA; (A5'UTR) 5'UTRs completely or partially antisense to a protein-coding region; (3'UTR) 3'untranslated region of an mRNA transcript; (A3'UTR) 3'UTRs completely or partially antisense to a protein-coding region.