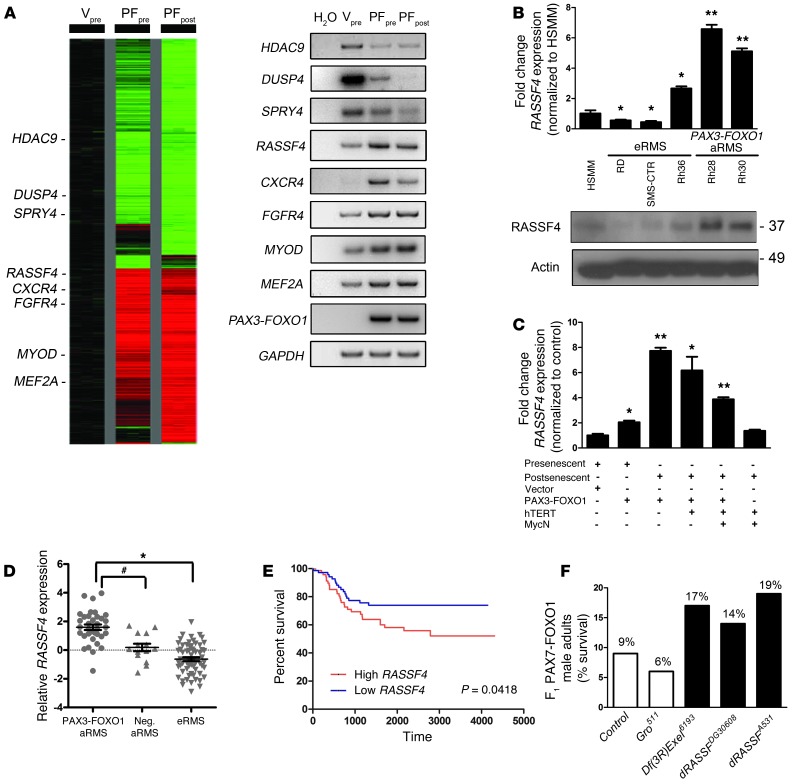
Figure 1. RASSF4 is upregulated in PAX3-FOXO1–positive aRMS cells and tumors.
(A) Left: Expression profile of HSMM control cells (Vpre) compared with PAX3-FOXO1–expressing HSMM presenescent (PFpre) or postsenescent (PFpost) cells. Right: Semiquantitative RT-PCR validation of select genes identified in the microarray. (B) PAX3-FOXO1–expressing aRMS cells expressed more RASSF4 than eRMS cells or HSMMs, as measured by qPCR and immunoblotting. *P < 0.05; **P < 0.005. Labels for cell lines correspond to qPCR and immunoblotting. Actin was used as a loading control. (C) HSMM-based model of aRMS displayed enhanced RASSF4 expression in a PAX3-FOXO1–dependent manner as measured by qPCR. *P < 0.05; **P < 0.005. (D) PAX3-FOXO1–positive primary human aRMS tumors expressed more RASSF4 than fusion-negative aRMS or eRMS. Error bars represent SEM. *P < 0.0001; #P = 0.0004; Mann-Whitney U test. Median-centered log2 values are shown, and microarray data were obtained from the Oncogenomics database. (E) RMS patient survival based on RASSF4 expression. The median RASSF4 expression value for RMS was the threshold for high versus low RASSF4 expression. High RASSF4, n = 73, Low RASSF4, n = 73. P value is based on log-rank test analysis. (F) dRASSF mutation genetically suppressed PAX7-FOXO1 pathogenicity in a Drosophila aRMS model. PAX7-FOXO1 expression in differentiating larval muscle causes semilethality, as PAX7-FOXO1 adults comprise only 9% of F1 adults (n = 170). (In Mendelian ratios, the F1 population should be composed of 50% wild-type and 50% PAX7-FOXO1 adults). The Df(3RExcel)6193 chromosomal deletion and dRASSFDG30608 and dRASSFA531 loss-of-function alleles suppressed PAX7-FOXO1–induced lethality. Groucho511 (Gro511) is an unrelated mutation included as a representative example of a nonsuppressor. Df(3R)Exel6193, n = 72; dRASSFA531, n = 130; dRASSFDG30608, n = 66; and Gro511, n = 116.