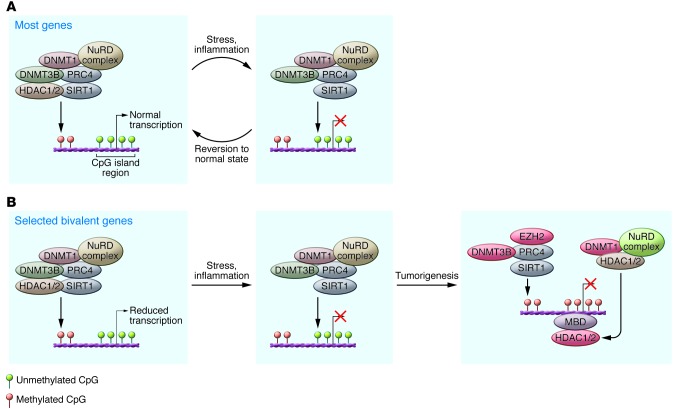
Figure 2. Model for initiation of DNA methylation-mediated gene silencing.
(A) Left panel: Methylation of CpGs at the borders of CpG islands in normal cells is mediated by the DNMTs as a complex with the HDACs, PRC4 complex (PRC2 component of PcG including SIRT1) with potential involvement of nucleosome remodelers (NuRD). In normal cells, the DNA methylation machinery is restricted from CpG islands, thus protecting these regions from methylation. Right panel: Movement of the repressive complex into the CpG island with stress and associated gene silencing, marked with a red X. However, most genes revert back to the normal cell state. (B) Left panel: A subset of genes with CpG island promoters in embryonic and adult stem cells have both the active (H3K4me3) and inactive (H3K27me3, PcG mark) marks at their promoters, termed “bivalent” marks, in which the genes have a low but poised expression state. Genes that are hypermethylated in cancers frequently have such bivalent marks in the embryonic and adult stem cells. When repetitive insults to cells, such as inflammation, recruit the DNA methylation machinery into the CpG islands of these vulnerable genes, this can initiate abnormal methylation for some of these genes (far right panel). The NuRD complex, a key platform for HDAC1 and -2, may be a part of the complex that maintains silencing of the methylated CpG islands containing genes. Various components of the epigenetic machinery that mediate gene silencing are being targeted already (red ovals), and others are potential targets (green ovals) for cancer therapy.