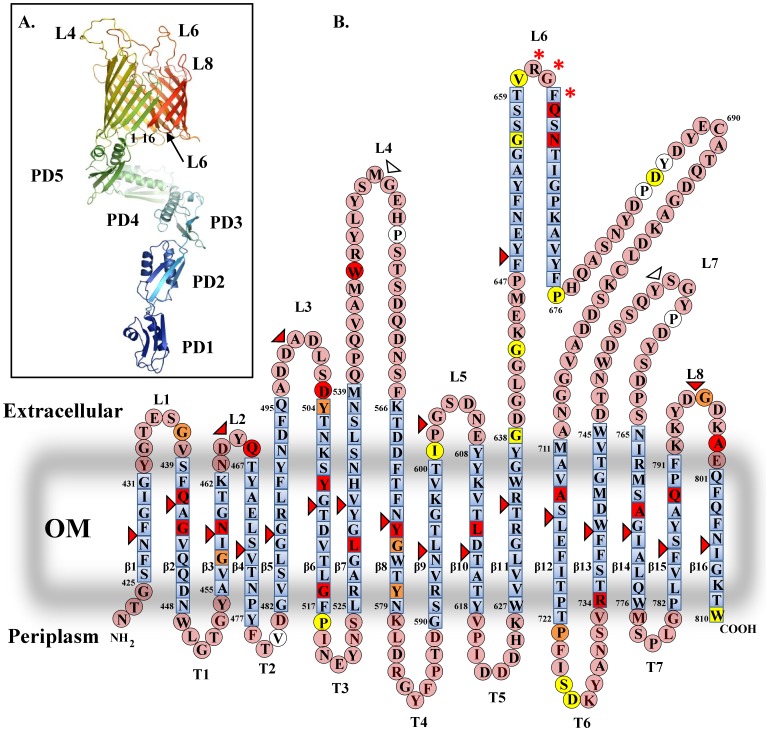
Figure 2. Topology model of the BamA β-barrel.
(A) The panel shows a model of BamA, displaying the five POTRA domains and β-barrel of BamA. The model was generated by combining the available crystal structures of BamA POTRA1 to POTRA4 (3ECF) [8] and POTRA4 and POTRA5 (3OG5) [10] with the model of the BamA β-barrel generated in this study using Coot [33] and is visualised using PyMol [35]. POTRA domains are indicated (PD1 to PD5), as are β-strands β1 and β16. The tip of loop L6 has been placed within the pore of the β-barrel lumen and has been modelled as a β-hairpin. (B) The panel shows a topology model of the BamA β-barrel (N422 to W810) derived from bioinformatics predictions. Amino acids within β-strand regions are shown as blue squares and those in external loops and periplasmic turns are shown as pink circles. β-strands β1 to β16, extracellular loops L1 to L8 and periplasmic turns T1 to T7 are indicated. The tip region of L6 is predicted to form a β-turn and the RGF motif important in BamA function is starred [14], [15]. The figure also details the position of 5 amino acid insertions isolated by linker scanning mutagenesis (Table S3). Insertions that either failed to rescue BamA depletion or did not allow growth in the presence of vancomycin (severe mutations) are coloured red, whilst insertion constructs which allowed growth on vancomycin concentration of 37.5, 75 and 150 µg ml−1 are coloured orange, yellow and white, respectively. The location of HA epitopes inserted within the β-barrel is also indicated using triangles (Table S5). The severity of the insertion is colour coded as for the linker scanning mutations.