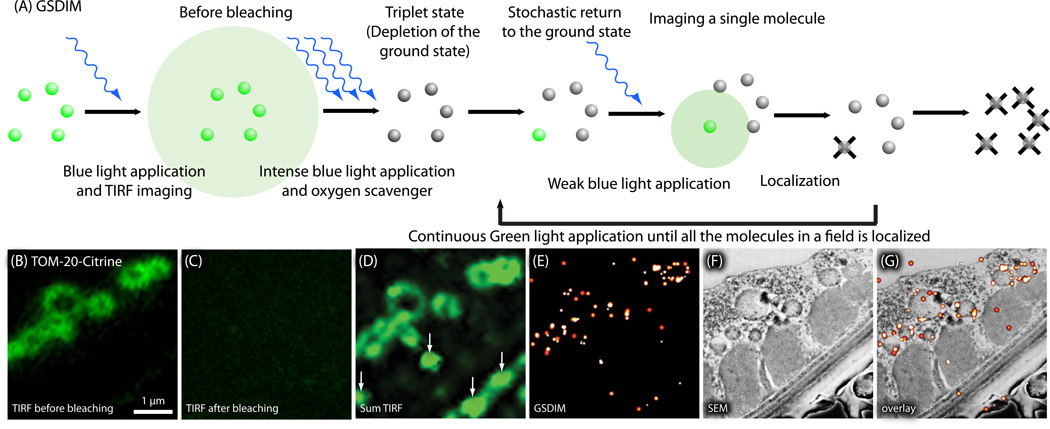
Fig. 8.
Nano-fEM using ground-state depletion (GSDIM). (A) Schematic diagram showing the concept of GSDIM. Fluorescent proteins, Citrine, are excited and driven into a dark state with intense laser in the presence of an oxygen scavenger. Fluorophores return to the ground state stochastically and are imaged. The centroid of each fluorescent spot is calculated and localized in a similar manner as PALM to reconstruct a final image. (B) TIRF image of TOM-20-Citrine acquired from a thin section (80 nm). Fluorescence signals are diffraction limited because all molecules fluoresce at the same time. (C) TIRF image of the same region imaged in (B). All the molecules are driven into a dark state (the triplet state) by intense 488 nm laser, and thus no signals are observed. (D) Sum TIRF image reconstructed from all the fluorescence spots detected by the camera. Background fluorescence is typically higher using GSDIM because sections cannot be pre-bleached using the readout laser. (E) Corresponding GSDIM image of TOM-20- Citrine. (F) Electron micrograph of a body wall muscle acquired from the same section. (G) Correlative GSDIM and electron microscopy of TOM-20-Citrine. The fluorescent signals are localized to the outer membrane of the mitochondria.