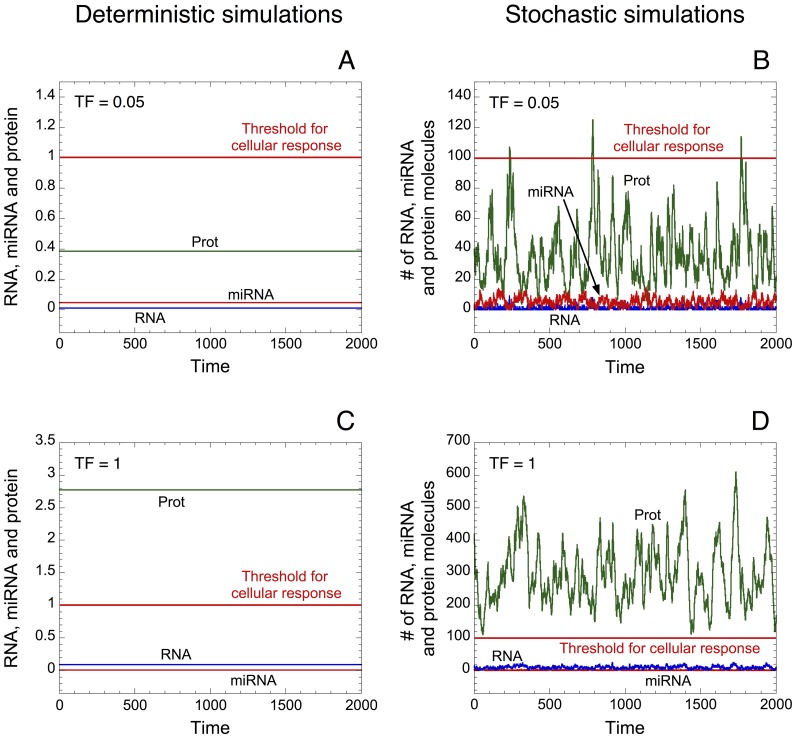
Figure 2. Effect of stochastic fluctuations on protein expression.
Deterministic (A, C) and the corresponding stochastic (B, D) time evolution of RNA, protein and miRNA are shown in the presence of low (TF = 0.05 in A, B) or high levels of TF (TF = 1 in C, D). The level of protein is low with small levels of TF and large with high levels of TF (compare panels A and C). Here again, horizontal red lines define an arbitrary threshold of protein needed to elicit a cellular response. Small stochastic fluctuations in the copy number of messenger RNA molecules can greatly affect the level of protein output (see panels B and D). Stochastic simulations were performed by means of the Gillespie's algorithm [87] using the stochastic version of the minimal model for protein synthesis (see Table 3). The units on the axes for the stochastic curves are expressed in numbers of molecules. The corresponding concentrations for the deterministic trajectories are obtained by dividing the numbers of molecules by Ω expressed in units of 106 L/NA, where NA is Avogadro's number. Here, as well as in Figs. 3 and S1, Ω = 100. The rate of synthesis of miRNA, V SMIRNA, is equal to 0.01; the coefficient of cooperativity, n, is equal to 1; and the other parameter values are as in Table 2.