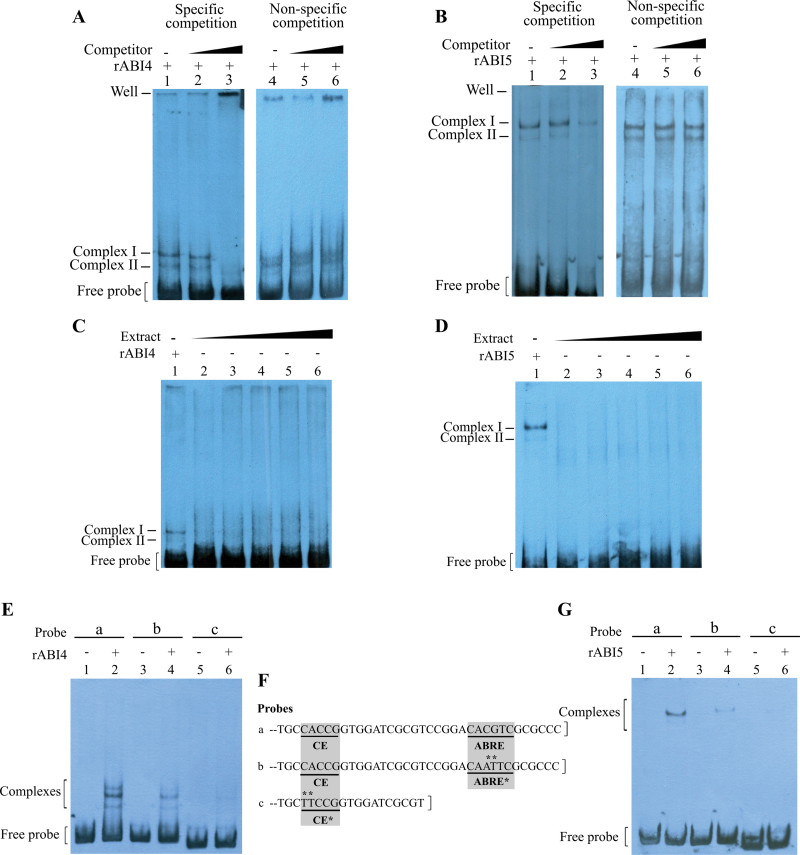
Fig. 3.
(A and B) Competition EMSAs for rABI4 and rABI5 performed with a 242bp SbGA2ox3 fragment (–505bp to –263bp) as biotinylated probe. For all lanes, 40ng of biotinylated probe and 0.225 μg of rABI4 (A) or 0.375 μg of rABI5 (B) were used. For specific competitions, the mass of unlabelled competitor DNA (SbGA2ox3 unlabelled fragment) in each reaction was as follows: lane 1, no competitor; lane 2, 200ng; lane 3, 600ng. For non-specific competitions, the mass of unlabelled competitor (SbGAMyb 183bp fragment) was as follows: lane 4, no competitor; lane 5, 200ng; lane 6, 600ng. (C and D) Control EMSAs for rABI4 (C) and rABI5 (D) performed with increasing concentrations of protein extract from E. coli cultures (transformed with empty pET24a) and 40ng of SbGA2ox3 biotinylated probe. For both C and D, the protein extract amount in each lane was as follows: lane 2, 10.74 μg; lane 3, 14.32 μg; lane 4, 17.9 μg; lane 5, 21.48 μg; lane 6, 25.06 μg. Lane 1 shows the positive control incubation reaction with 40ng of SbGA2ox3 biotinylated probe and 0.225 μg of rABI4 (C) and 0.375 μg of rABI5 (D). (E and G) EMSAs carried out with rABI4 (E) or rABI5 (G) and shorter SbGA2ox3 probes a, b, and c. For all lanes, 40ng of biotinylated probe was used. The protein amount was as follows: lanes 1, 3, and 5, no protein; lanes 2, 4, and 6, 0.225 μg of rABI4 (E) and 0.375 μg of rABI5 (G). (F) Sequences of probes a, b, and c with highlighted CEs and ABREs. Probe a, wild-type probe (intact ABRE and CE); probe b, mutated ABRE; probe c, mutated CE and no ABRE included. Asterisks in probes b and c indicate mutated bases and elements. Probe size was 131bp for a and b, and 114bp for c. (This figure is available in colour at JXB online.)