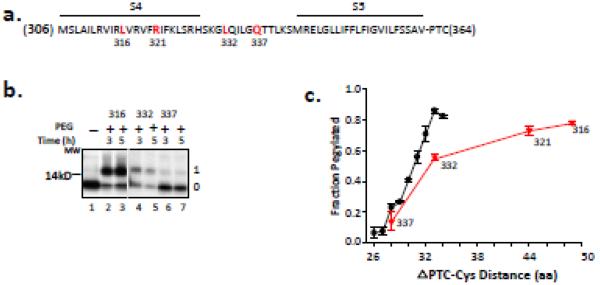
Figure 2. Accessibility of S4 residues.
(a) Primary sequence of S4 and flanking residues. The amino acid sequence is indicated by a single-letter code. Residues 316, 321, 332 and 337 (bolded, red) were mutated to cysteines, one at a time, and used in the pegylation assay. The N-terminus of the peptide is M306 and a restriction enzyme (Acc1) was used to generate the terminal amino acid, V364, at the peptide’s C-terminus. The peptide thus includes S4-S5, attached to the PTC at residue 364. (b) Pegylation of selected S4 residues of Kv1.3 indicated in (a). Nascent peptides were translated from mRNA engineered from DNA that lacked a stop codon, pegylated (1 mM PEG-MAL) for the times indicated, and fractionated using SDS-PAGE as described previously4. The unpegylated protein is shown in lane 1 and is identical for all peptides substituted with cysteines at 316, 332, and 337. Gels were 12% NuPAGE Bis-Tris gels with MES running buffer. The number to the left of the gel is a molecular weight standard; numbers to the right of the gel indicate unpegylated (0) and singly pegylated (1) protein. (c) Fraction pegylated for the S4 residues (red triangles) indicated along the x-axis, which indicates the number of amino acids from the PTC to (and including) the labeled cysteine. Pegylation of a known all-extended nascent peptide is shown by the black circles and represents data taken from Lu and Deutsch, 20052. Symbols are mean ± SEM (n= 3).