Figure 1.
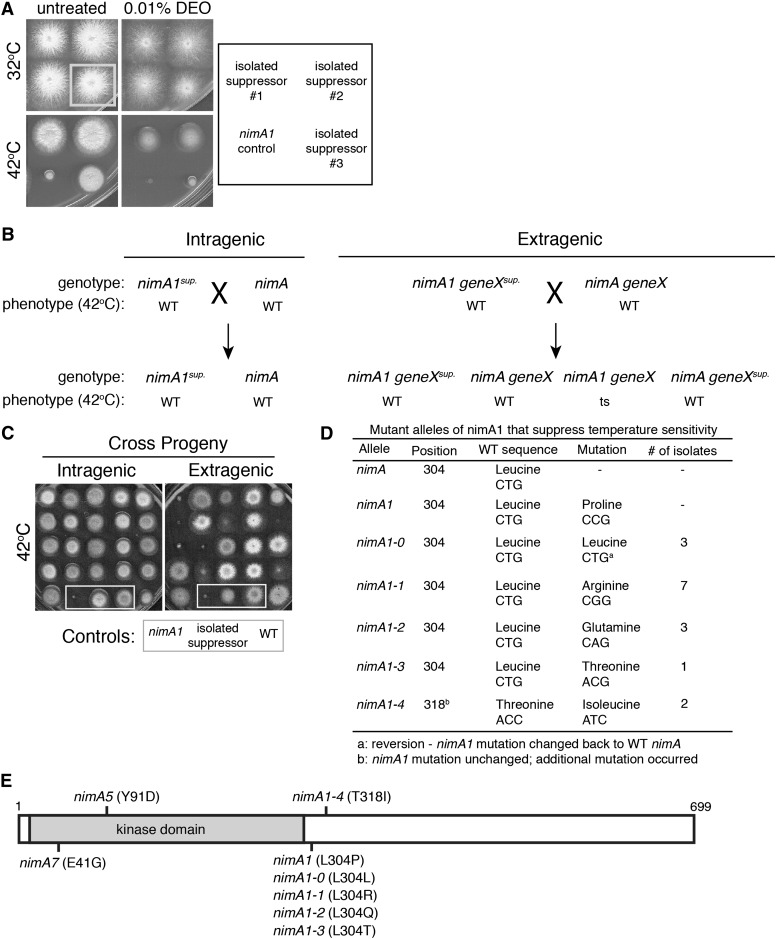
Characterization of intragenic nimA1 suppressors identified new alleles of nimA. (A) Three different nimA1 suppressors isolated from the screen were replica-plated onto plates with or without 0.01% DEO and incubated at 32° or 42° for 2 days. (Bottom left) The unmutagenized nimA1 control strain (JLA1) was spotted for comparison. Although all three mutants suppressed the temperature sensitivity of nimA1, only one of them (#3) displayed temperature-dependent DNA damage sensitivity. (B) Diagram illustrating how intragenic suppressors were identified. Shown are the expected outcomes from crosses between isolated nimA1 suppressors and a wild-type (WT) strain to distinguish intragenic from extragenic suppressors. (C) The temperature sensitivity caused by nimA1 segregates independently from extragenic suppressors. Plates grown at 42° for 2 days show examples of progeny from crosses diagrammed in B. Extragenic suppressors were identified based on the isolation of temperature-sensitive progeny from a cross with a wild-type strain. In contrast, all progeny from a cross between a wild-type strain (C61) and intragenic suppressors (mutagenized strain JLA1) are not temperature-sensitive. Control strains are boxed by a shaded outline. Strain JLA1 was used as the nimA1 control. (D) Table of new intragenic NIMA alleles isolated as suppressors of nimA1 temperature sensitivity. (E) Diagram of NIMA with the positions of identified alleles marked. The catalytic domain is shaded.