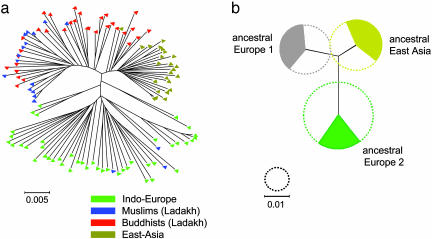
Fig. 3.
A multilocus haplotype tree and quantitative sources of nucleotides from three ancestral populations for 119 H. pylori isolates from Ladakh, East Asia, and Indo-Europe. (a) Neighbor-joining tree of individual haplotypes (Kimura two-parameter model). Fig. 6, which is published as supporting information on the PNAS web site, includes the designation of each individual isolate. (b) Sources of nucleotides. The diameters of the three circles indicate the genetic diversity of the three ancestral populations and the filled arcs indicate the proportion of the nucleotides in the 119 haplotypes that are derived from each of these populations. The black lines joining the ancestral populations represent a neighbor-joining population tree as measured by 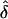 , the net nucleotide distance between populations.
, the net nucleotide distance between populations.