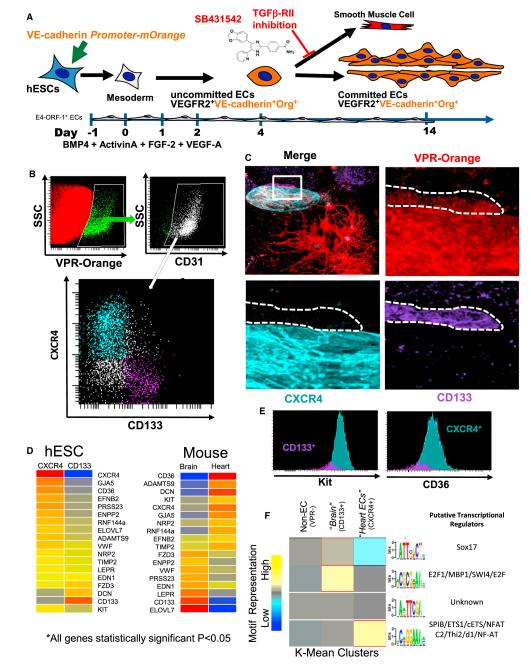
Figure 6. ECs Derived from hESCs Phenocopy Adult Mouse Tissue-Specific Capillaries.
(A) Schema of in vitro conditions to support the differentiation and identification of hESC-derived vasculature. hESCs are grown on an E4-ORF1 EC feeder layer and transduced with a VE-Cadherin-Orange reporter gene. VE-Cadherin-Orange+ vascular networks are readily identifiable by day 10.
(B) Flow cytometry data depicting the expression of VPR-Orange on hESC-derived CD31+ ECs. These VPR+ ECs have distinct populations based on the expression of either CXCR4 (teal) or CD133 (purple).
(C) VPR+CXCR4+CD133− and VPR+CD133+CXCR4− ECs are capable of forming distinct clusters of ECs in hESC cultures.
(D) Heat maps of the genes, which were common in their statistically significant differential expression (Benjamini-Hochberg adjusted p < 0.05) between hESC-derived vasculature and adult mouse heart and brain tissues.
(E) VPR+CXCR4+CD133− and VPR+CD133+CXCR4− ECs were analyzed for cKit and CD36 levels via flow cytometry. Validation of the higher expression of CD36 and Kit in the CXCR4+ ECs is shown.
(F) Heat map of K-Mean clusters depicting the results of de novo motif discovery among non-ECs, CXCR4+VPR+ ECs, and CD133+VPR+ ECs. Candidate binding partners to the motifs are listed.