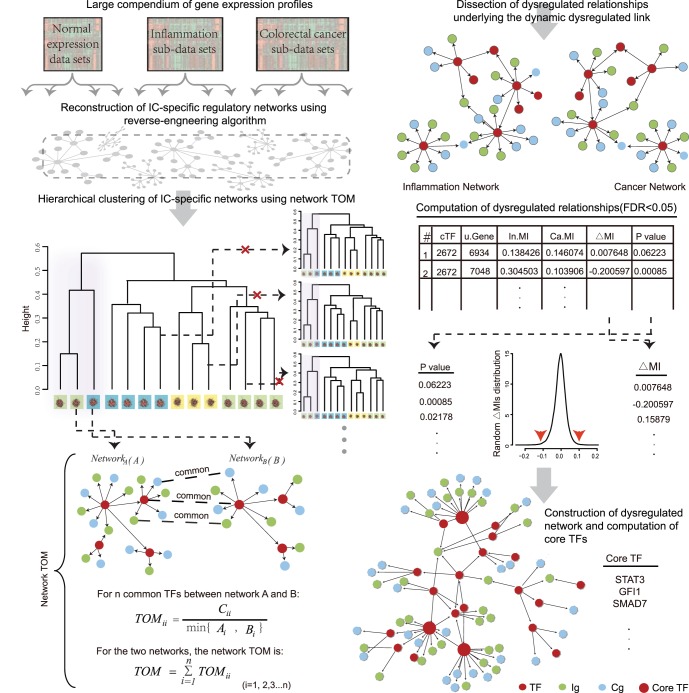
Figure 1. Workflow applied to identify core TFs.
The procedure is mainly divided into four steps: 1) reconstruction of IC-specific regulatory networks from a large compendium of microarray data; 2) clustering of IC-specific regulatory networks using network TOM, followed by network perturbations; 3) construction of a dysregulated network with edges dysregulated between inflammation and their linked cancer network based on network comparisons; 4) identification of core TFs via dysregulated pattern analysis. TF, transcription factor; Ig, inflammation gene; Cg, colorectal cancer gene; MI, mutual information.