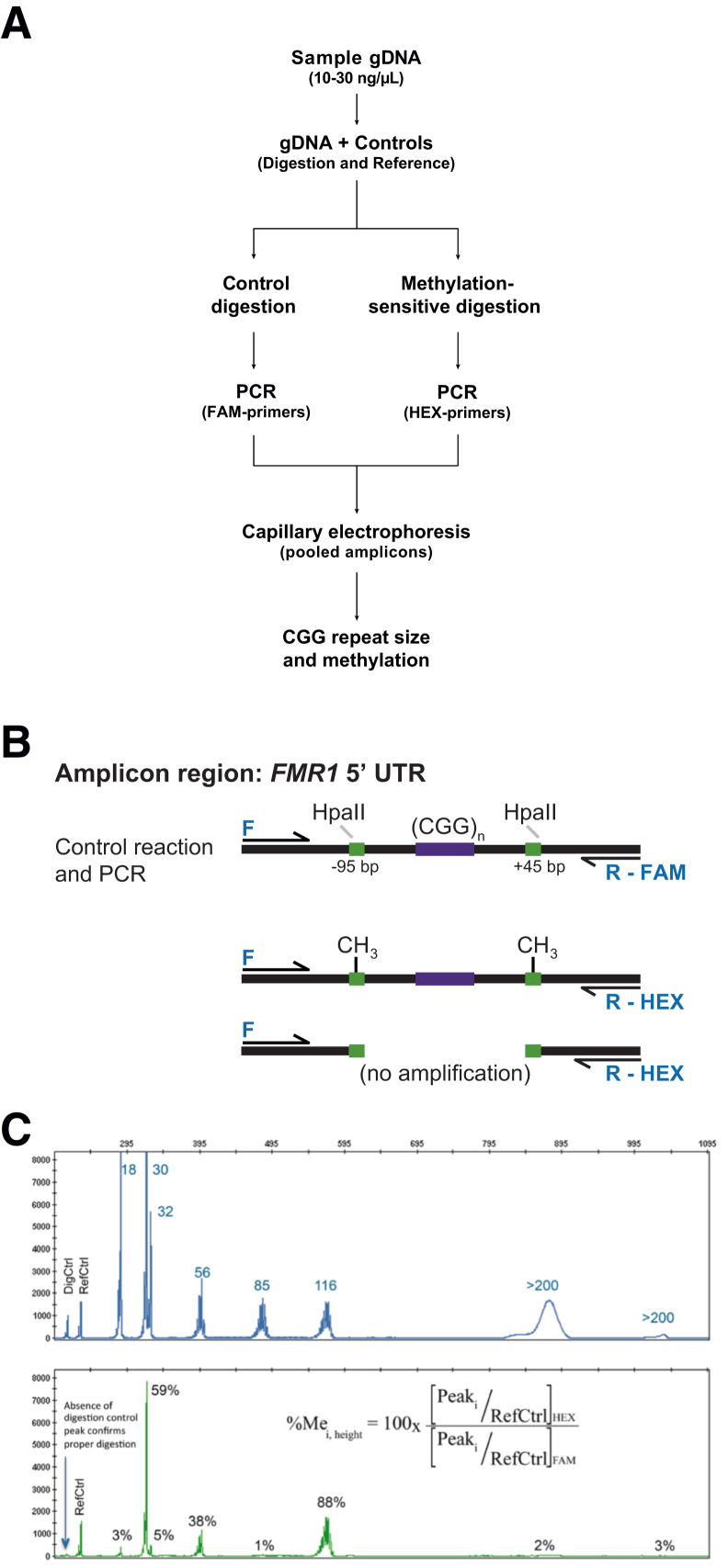
Figure 1.
A: Methylation PCR (mPCR) workflow. DNA samples were prepared for methylation assessment by mixing with procedural controls and treating with separate methylation-sensitive restriction and control reactions followed by PCR using different dye labels. The HEX- and FAM-labeled amplicons were pooled and sized using capillary electrophoresis. B: Schematic representation of the amplicon region and outcomes of digestion and PCR. The PCR primers spanned two HpaII restriction sites 95 bp from the beginning and another 45 bp from the end of the repeat region using a forward (F) primer and either a FAM- or HEX-labeled reverse primer: R-FAM or R-HEX, respectively. Products from the control digestion were intact and amplified using FAM-labeled primers, but after digestion with HpaII, only alleles fully methylated at both sites were amplified using HEX-labeled primers. Lack of methylation at either site resulted in no amplification. C: Example of two-color data profiles for a multi-allele control. Electropherograms in the FAM channel yielded the total FMR1 profile for that sample and determination of CGG repeats. The signal intensity in the HEX channel corresponded to the methylated component of that allele amplicon. The percent methylation was determined using the ratio of peak heights for each in both channels (equation) normalized to the peak height of the reference control (RefCtrl), one of the plasmid DNA controls spiked into each PCR. Reduction of signal in the HEX channel for the digestion control (DigCtrl), another plasmid DNA spiked into each reaction, correlated with activity of HpaII with expected reduction of signal in the HEX channel. gDNA, genomic DNA; UTR, untranslated region.