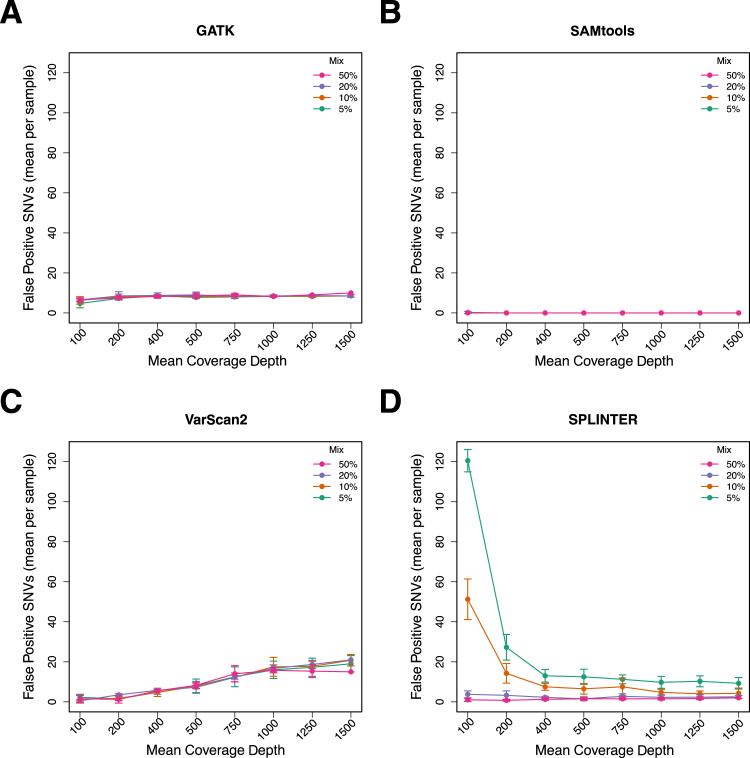
Figure 4.
False positives (mean per sample) called by GATK, SAMtools, VarScan2, and SPLINTER as a function of mean target coverage and mix proportion across the entire target region encompassing coding and noncoding sequences of 26 genes (306,336 bp). Variant SNV calls in down-sampled data that were not among the gold standard SNVs were called as false positive, after excluding indel calls and positions that were low coverage (<50) or discordant in the gold standard analysis. The number of false positive calls for GATK (A), SAMtools (B), VarScan2 (C), and SPLINTER (D) for each down-sampled coverage level and the mix proportion indicated by the legend. Error bars show the SD for each coverage level and mix proportion.