FIGURE 11.
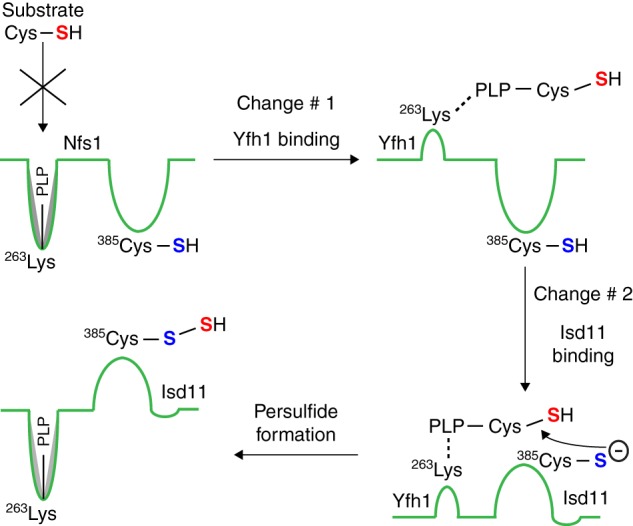
Model for two-tier regulation of persulfide formation by Nfs1. The enzyme Nfs1 is shown by a green line. The Lys263 residue of the Nfs1 substrate-binding site forms a Schiff base with the cofactor PLP. Nfs1 by itself inefficiently binds the substrate cysteine (Cys-SH, sulfur indicated in red). Frataxin/Yfh1 interacts with Nfs1, inducing a conformational change in the enzyme (Change #1) and exposing new sites with Lys263-PLP, now able to efficiently bind the substrate cysteine. Isd11 then interacts with Nfs1 with substrate cysteine already bound, inducing a second conformational change (Change #2), and bringing the substrate cysteine (red sulfur) into proximity to the Cys385 (sulfur indicated in blue) in the peptide backbone of the Nfs1 active site loop. A nucleophilic attack by the thiolate anion of the active site Cys385 extracts the –SH group of the substrate, forming a covalent persulfide (Nfs1-S-SH). The substrate cysteine bound to PLP after Change #1 is shown as an external aldimine. However, this has not been shown experimentally, and it is possible that at this stage the substrate cysteine interacts with PLP still bound to the Lys263 residue of Nfs1 as an internal aldimine.
