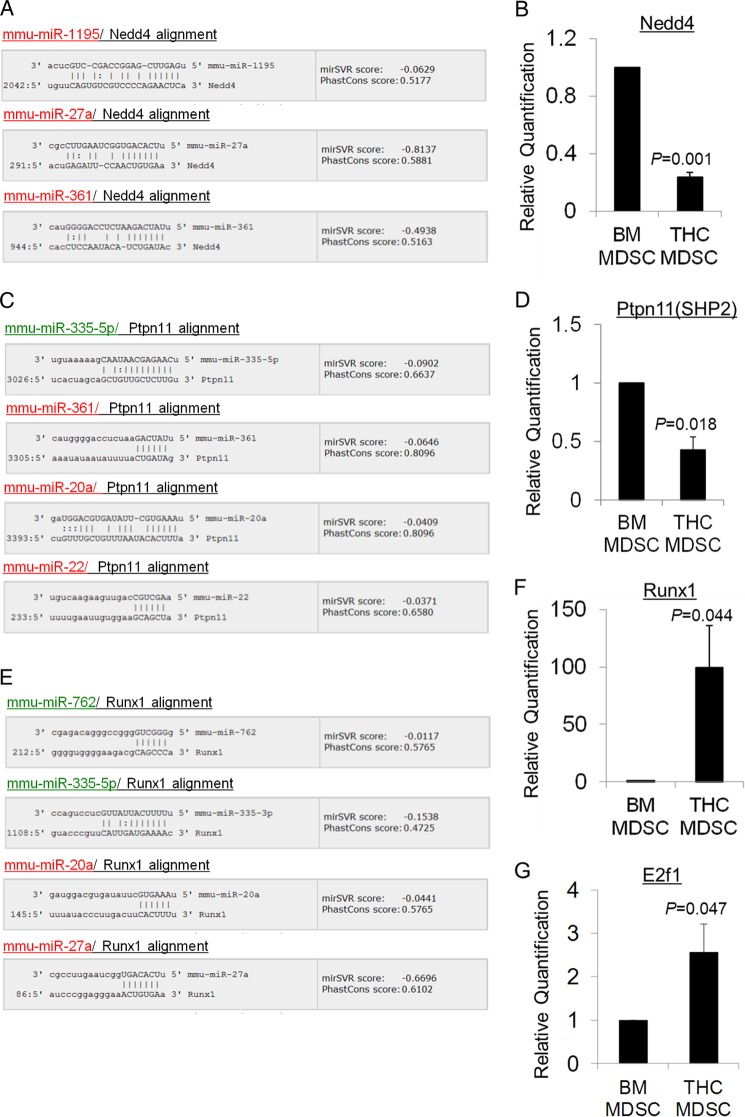
FIGURE 8.
Experimental validation of in silico predicted miRNA target genes. Some of the predicted target genes of differentially expressed miRNA in THC-MDSC relative to BM precursors involved in top enriched pathways were validated by qPCR. miRanda-generated alignments of target binding sites on 3′-UTR of Nedd4, Ptpn11, and Runx1 with seed sequence of differentially expressed miRNA with good mirSVR scores are shown (A, C, and E). 6-mer or better seed match and mirSVR score cutoff of ≤−0.01 was used, and the top four predictions are shown for each. mirSVR down-regulation scores represent the likelihood of target mRNA down-regulation (log fold change) for each miRNA-mRNA predicted target site and are based on a regression model that takes into account sequence match as well as structure-stability features and is calibrated to correlate linearly with the extent of down-regulation. PhastCons conservation scores for the UTR target sites are also shown. Up-regulated miRNA are highlighted in red, and down-regulated miRNA are in green. The results of real time qPCR analysis of gene transcripts normalized to 18 S in purified control BM precursors and THC-induced MDSCs are depicted in B, D, F, and G.