FIGURE 2.
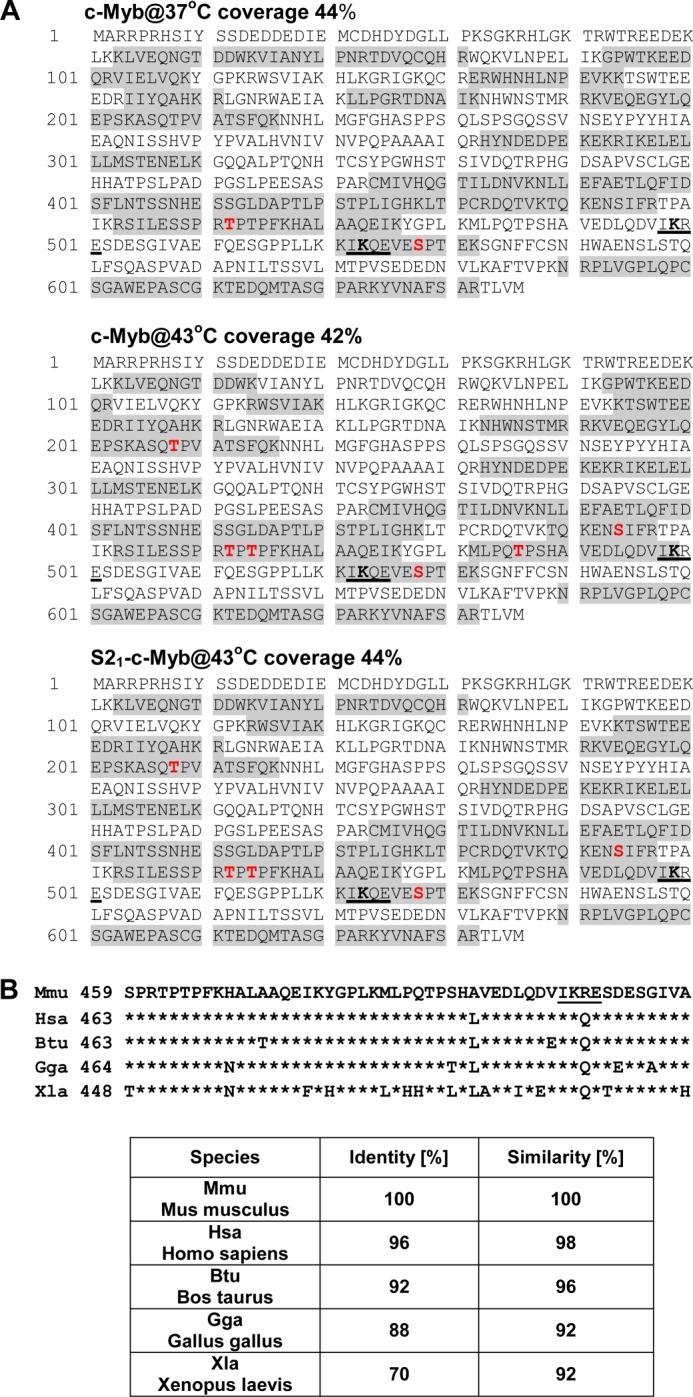
Identification of phosphorylation sites in c-Myb isolated from stressed cells by mass spectrometry. A, protein sequence of mouse c-Myb protein. The shaded residues were identified in the tryptic peptides from the nonSUMOylated form of c-Myb isolated from untreated cells (c-Myb@37°C) and nonSUMOylated c-Myb (c-Myb@43°C) and SUMOylated c-Myb (S21-c-Myb@43°C) from cells treated with hyperthermia. Total protein coverage is shown for every sample analyzed. Phosphorylation sites identified in analyzed samples are marked in red. Three new phosphorylation sites Thr208, Ser444, and Thr486 were identified in c-Myb isolated from stressed cells. SUMOylation motifs 498IKRE501 and 522IKQE525 are underlined. B, conservation of c-Myb protein sequences (459–509 amino acids, murine numbering). Phosphorylation site Thr486 is located amino-terminally to SUMOylation site Lys499. The SUMOylation motif IKRE is underlined. Identity and similarity of the c-Myb sequences between mouse and other species are presented in the table.
