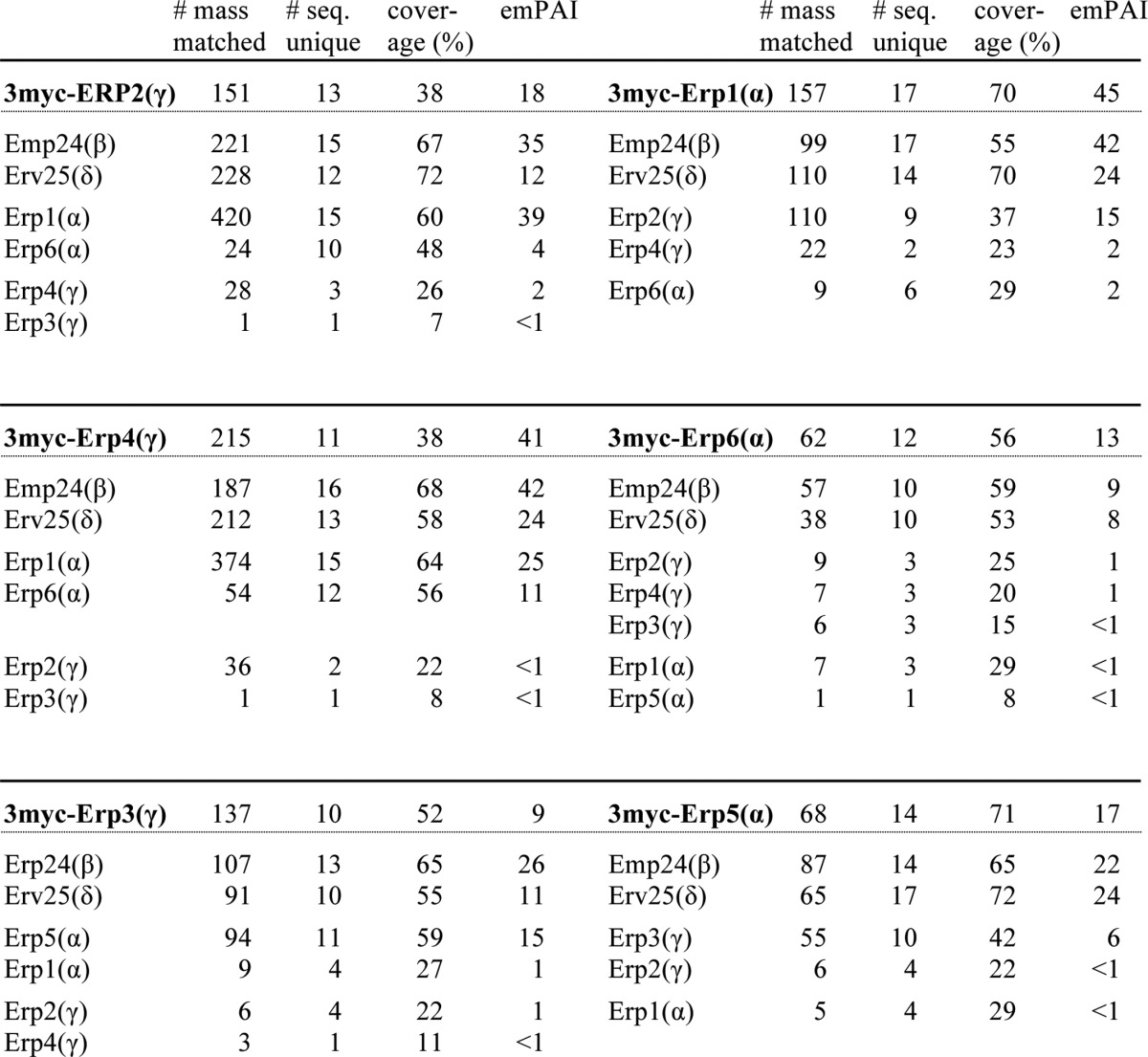
TABLE 4.
Mass spectrometry of p24 immunoprecipitates
3 myc-p24 immunoprecipitates were resolved by SDS-PAGE. Gel regions expected to contain proteins of 15–37 kDa were excised and subjected to mass spectrometry. Total number of mass matched, the number of sequences uniquely assigned to the proteins of interest, sequence coverage, and emPAI are shown. emPAI (exponentially modified protein abundance index), which offers approximate and relative quantification of the proteins in a mixture, was calculated on two parameters, the number of experimentally observed peptides as in Ishihama et al. (79). The emPAI values for p24α and -β (Emp24) were calculated to be higher than those for p24γ and -δ (Erv25). We speculate this reflects ionization efficiency of these proteins rather than their stoichiometry in the complex.