FIGURE 1.
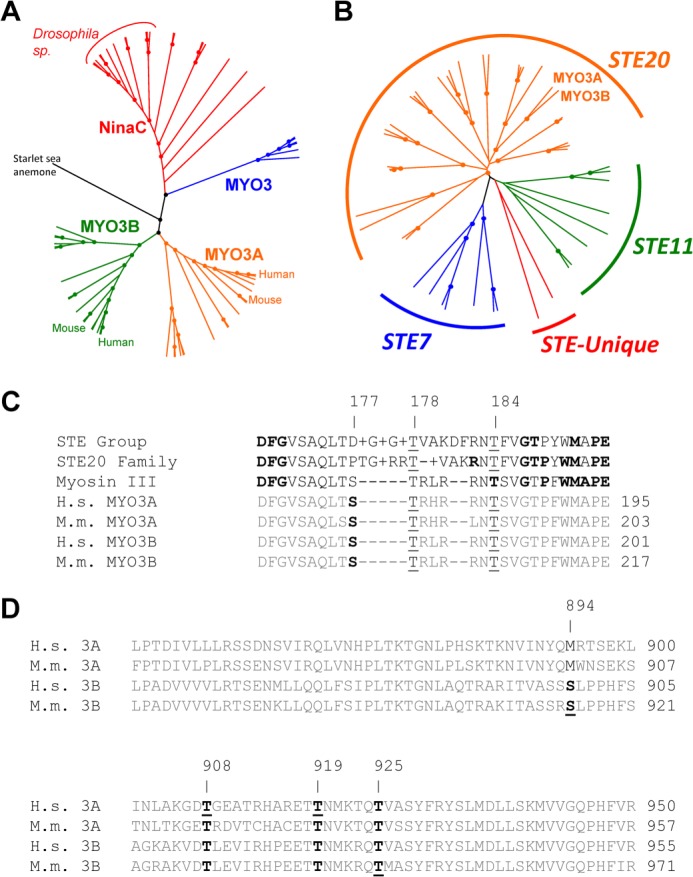
Sequence analysis of the MYO3A kinase domain. Unrooted phylogenetic trees of 61 class III myosins (A) and STE group kinases in the human genome (B) generated from ClustalW2 (default settings) alignments. Dots represent nodes validated by bootstrapping values of greater than 80%. C, alignments of the activation loop of MYO3A with consensus sequences from the STE group, STE20 family, and class III myosins. Boldface residues in the STE group, STE20 family, and class III consensus sequences represent 80% conservation at that position within that category. The underlines indicate well conserved phosphorylatable residues corresponding to positions 177, 178, and 184 of human MYO3A. Although threonine is the consensus residue at the 184 position, it is not completely conserved. However, all sequences except for MAP3K1 and COT contain a phosphorylatable residue within 1 amino acid of the 184 position. D, alignments of the human and mouse MYO3A and MYO3B loop 2 sequences from within the myosin motor domain. Boldface residues indicate well conserved phosphorylatable sequences in either MYO3A or MYO3B. Underlined residues indicate identified phosphorylation sites in human MYO3A from this study, and mouse MYO3A and MYO3B from a previous study (21).
