Background: Sterol 12α-hydroxylase catalyzes cholic acid synthesis. RORα is a cholesterol-activated and fasting-induced nuclear receptor and core clock gene.
Results: Upon fasting, glucagon/PKA phosphorylated and stabilized RORα protein to induce CYP8B1 and diurnal rhythm.
Conclusion: RORα is a key regulator of CYP8B1 that regulates bile acid and cholesterol homeostasis.
Significance: Antagonizing RORα activity may be a therapeutic strategy for treating non-alcoholic fatty liver disease and diabetes.
Keywords: Cholesterol, Diabetes, Lipid Metabolism, Nuclear Receptors, Obesity
Abstract
Sterol 12α-hydroxylase (CYP8B1) is required for cholic acid synthesis and plays a critical role in intestinal cholesterol absorption and pathogenesis of cholesterol gallstone, dyslipidemia, and diabetes. In this study we investigated the underlying mechanism of fasting induction and circadian rhythm of CYP8B1 by a cholesterol-activated nuclear receptor and core clock gene retinoic acid-related orphan receptor α (RORα). Fasting stimulated, whereas restricted-feeding reduced expression of CYP8B1 mRNA and protein. However, fasting and feeding had little effect on the diurnal rhythm of RORα mRNA expression, but fasting increased RORα protein levels by cAMP-activated protein kinase A-mediated phosphorylation and stabilization of the protein. Adenovirus-mediated gene transduction of RORα to mice strongly induced CYP8B1 expression, and increased liver cholesterol and 12α-hydroxylated bile acids in the bile acid pool and serum. A reporter assay identified a functional RORα response element in the CYP8B1 promoter. RORα recruited cAMP response element-binding protein-binding protein (CBP) to stimulate histone acetylation on the CYP8B1 gene promoter. In conclusion, RORα is a key regulator of diurnal rhythm and fasting induction of CYP8B1, which regulates bile acid composition and serum and liver cholesterol levels. Antagonizing RORα activity may be a therapeutic strategy for treating inflammatory diseases such as non-alcoholic fatty liver disease and type 2 diabetes.
Introduction
Bile acids are synthesized from cholesterol in the liver. Cholic acid (CA)2 and chenodeoxycholic acid (CDCA) are two primary bile acids synthesized in human liver. Bile acids are conjugated to glycine or taurine and secreted into bile and stored in the gallbladder. After each meal bile acids are secreted into the digestive tract. In the intestine, CA and CDCA are converted to the secondary bile acids, deoxycholic acid and lithocholic acid, respectively, by 7α-dehydroxylase activity in bacterial flora. About 95% of bile acids are reabsorbed in the ileum and circulated back to the liver via portal blood to inhibit bile acid synthesis. Enterohepatic circulation of bile acids plays a critical role in absorption, distribution, metabolism, and disposal of nutrients, drugs, xenobiotics, and endobiotics (1). Bile acids are endogenous ligands that activate the nuclear receptors farnesoid X receptor (FXR), pregnane X receptor, and vitamin D receptor (1) and also activate the membrane G protein-coupled receptor TGR5 to maintain glucose, lipid, and energy homeostasis and to protect against inflammation in liver and intestine (2–4). The rate of bile acid synthesis is determined by cholesterol 7α-hydroxylase (CYP7A1), whereas the ratio of CA to CDCA in bile is determined by sterol 12α-hydroxylase (CYP8B1) in CA synthesis (1). CA forms mixed micelles with cholesterol and phospholipids at low critical micelle concentrations for storage in the gallbladder and to efficiently facilitate intestinal absorption of dietary cholesterol and fats (5). For that reason, CA (0.5%) is included in lithogenic diets to accelerate hypercholesterolemia, cholesterol gallstones, and atherosclerosis in C57BL6 mice (5, 6). Ablation of the CYP8B1 gene reduces dietary cholesterol absorption and prevents hypercholesterolemia and gallstone formation in diabetic mice (7–9) and atherosclerosis in ApoE knock-out mice (10). It has been reported that CYP8B1 expression is elevated in streptozocin-induced diabetic rats, and insulin inhibits CYP8B1 and regulates circadian rhythm of CYP8B1 (11).
Bile acid synthesis and CYP7A1 expression exhibit a strong circadian rhythm, peaking at the onset of dark and decreasing in the light cycle (12–15). It has been reported that peroxisome proliferator-activated receptor α, DEC1/2, and E4BP4 are negative regulators, whereas Rev-erbα and D-site binding protein are positive regulators of circadian rhythm of CYP7A1 (16, 17). Rev-erbα is a negative clock gene that stimulates CYP7A1 via inhibiting SHP (small heterodimer partner) and E4BP4, the negative regulators of CYP7A1 (17). In Clock gene mutant mice, the circadian rhythm of CYP7A1 is enhanced, but that of CYP8B1 is diminished (16). Two core clock genes and nuclear receptors, RORα and Rev-erbα, are known to play a key role in induction of the Clock/BMAL1 heterodimer in the positive limb to activate the Per/Cry dimer, which composes the feedback loop that inhibits BMAL1/Clock transcription (18). How clock genes regulate CYP8B1 expression remains to be determined.
Retinoic acid-related orphan nuclear receptor α (RORα, NR1F1) is a constitutively active transcriptional activator that plays various roles in regulation of cholesterol metabolism, circadian rhythm, bone formation, and inflammatory response (18–20). The ROR subfamily of nuclear receptors is expressed in many tissues including brown adipocytes, liver, and kidney and binds as a monomer to ROR-response elements (RORE) in target gene promoters. A natural mutation at the RORα allele has been identified in staggerer (sg/sg) mice (19). Microarray gene profiling of RORα and RORβ double knock-out mouse livers revealed reduced expression of several genes in phase I and phase II drug, steroid, and bile acid metabolism including CYP8B1 and oxysterol 7α-hydroxylase (CYP7B1) (21). It has been reported that RORα binds to and activates mouse Cyp7b1 gene transcription (22), but the role of RORα in regulation of CYP8B1 has not been studied. The ligand binding pocket of RORα contains cholesterol or cholesterol sulfate, which may be endogenous RORα ligands (23, 24). A recent study shows that 7α-oxygenated cholesterols are high affinity ligands for RORα and RORγ and that 7α-hydroxycholesterol (7α-OHC), a product of CYP7A1, is an inverse agonist of RORα that decreases RORα transcriptional activation of gluconeogenic genes to reduce hepatic glucose output (25). This could be a mechanism linking bile acid synthesis to inhibit gluconeogenesis. Interestingly, on a high fat diet sg/sg mice have reduced adiposity, inflammation, and hepatic steatosis and improved insulin sensitivity (19, 20).
It is thought that bile acids, via enterohepatic circulation, feedback-inhibit CYP8B1 gene transcription mainly by activating the FXR-small heterodimer partner (SHP) pathway (1). We reported recently that bile acid synthesis is regulated by the fasting-to-refeeding cycle. Glucagon inhibits, whereas glucose and insulin stimulate CYP7A1 gene transcription (11, 26, 27). In contrast, fasting stimulates, whereas feeding inhibits CYP8B1 gene expression and activity. The opposing effects on nutrient regulation of these two important regulatory genes in bile acid synthesis may alter bile acid composition, which affects hepatic lipid and glucose metabolism during fasting and re-feeding cycles (1). The molecular mechanism of regulation of CYP8B1 gene transcription by fasting and circadian rhythm is not known. In this study we explored the role of RORα in mediating fasting and diurnal regulation of CYP8B1 gene expression. Our results show that RORα is a key regulator of diurnal rhythm and fasting induction of the CYP8B1 gene. Fasting induced glucagon/cAMP/PKA signaling stimulates RORα activity by increasing RORα protein stability. Regulation of CYP8B1 is linked to pathogenesis of obesity-related inflammation, non-alcoholic fatty liver disease and diabetes.
EXPERIMENTAL PROCEDURES
Reporter Plasmids
Human CYP8B1/Luc reporter construct (nucleotide −514 to +300 CYP8B1/Luc) was constructed previously (28). Site-directed mutagenesis of the RORα response element on the human CYP8B1 reporter plasmid was conducted using a QuikChange site-directed mutagenesis kit (Agilent Technology, Santa Clara, CA). Human RORα expression plasmid was kindly provided by Dr. Wen Xie (University of Pittsburgh). RORα-VP16 plasmid was cloned by inserting RORα cDNA between the KpnI and BamH1 sites of the VP16 plasmid (Promega Corp.). Expression plasmids for CBP and Gal4-CBP were gifts from Dr. Carolyn Smith (Baylor College of Medicine, Houston, TX). Plasmids for peroxisome proliferator-activated receptor γ co-activator 1α (PGC1α), Gal4-PGC1α, Gal4-steroid receptor co-activator (SRC1), and Gal4-SRC2 were generated as reported previously (29).
Animals
Male wild type C57BL/6J mice were purchased from The Jackson Laboratory (Bar Harbor, ME). All mice were maintained on a standard chow diet and water ad libitum and housed in a room with a 12-h light (6 a.m. to 6 p.m.) and 12-h dark (6 p.m. to 6 a.m.) cycle; lights on at Zeitgeber time 0 (ZT0) and lights off at ZT12. Mice were fasted for 12 h (ZT2 to ZT14). A group of 4 mice was sacrificed at ZT14 as the “fasted” group, and the other group (n = 4) was refed at ZT14 for 6 h and sacrificed at ZT20 as “fed” group. To study the effect of restricted feeding on circadian expression of CYP8B1 and RORα, 24 mice were allow free access to food (free-feeding), and 24 mice were fasted 16 h from 8 a.m. (ZT2) to midnight (ZT18) and were refed with a standard laboratory chow diet for 3 h (from ZT18 to ZT21 (restricted feeding)). Four mice in each group were sacrificed at ZT2, -6, -10, -14, -18, and -22. Liver mRNA was isolated for analysis of CYP8B1 and RORα mRNA levels, and liver extracts were used for immunoblot analysis of CYP8B1 and RORα protein levels. The institutional animal care and use committee of Northeast Ohio Medical University approved all animal protocols used in this study.
Recombinant Adenovirus
Adenovirus carrying RORα was cloned using an Adeasy adenovirus vector system according to the manufacturer's protocol (Agilent Technology, Santa Clara, CA). Adenoviruses were purified from HEK293A cells by CsCl density centrifugation. Viral titers were determined by an Adeno-X Rapid Titer kit (Agilent Technology). RORα adenovirus (Ad-RORα) and adenovirus null (Ad-null) were administered at 2 × 109 pfu/mouse via tail vein injection (n = 4). Experiments were carried out 7 days post injection.
Cell Culture
Human hepatoblastoma HepG2 cells (American Type Culture Collection, Manassas, VA) were cultured as described previously (30). Primary human hepatocytes (donors 1839, 1859, and 1858) were obtained from the Liver Tissue and Cells Distribution System of National Institutes of Health (University of Pittsburgh, Pittsburgh, PA). Cells were maintained in hepatocyte maintenance medium as described previously (31).
RNA Isolation and Quantitative Real-time PCR
Total RNA was isolated with TRI Reagent (Sigma). All primers/probe sets for real-time PCR were ordered from TaqMan Gene Expression Assays (Applied Biosystems, Foster City, CA). For assay of RORα and GAPDH mRNA, SYBR Green primers (Applied Biosystems) were used as reported previously (29). Amplification of ubiquitin C and GAPDH were used as internal controls. Relative mRNA expression was quantified using the comparative CT (Ct) method and expressed as 2−ΔΔCt.
Electrophoretic Mobility Shift Assay (EMSA)
Probes were designed, purchased, and radiolabeled as described previously (30). RORα protein was generated by a transcription and translation lysate kit (TNT; Promega Scientific Inc., Madison, WI). EMSA assay was performed as described previously (30). The sequences of probes used were: APOCIII, −267gggatccgatataaaacaggtcagaaccccta−235; CYP8b1 (wt), +9ggagagctgtctgaaaaggtcagagcaaagca+39; CYP8b1-Mutant 1(M1), +9ggagagctgtctggggaggtcagagcaaagca+39; CYP8b1-Mutant 2 (M2), +9ggagagctgtctgaaaaccccagagcaaagca+39. The bold types indicate the putative RORα binding sites in APOCIII and CYP8B1 promoter. Italics indicate mutated nucleotides in RORα binding site in CYP8B1 promoter.
Luciferase Reporter Assay
HepG2 cells were grown to about 80% confluence in 24-well tissue culture plates and treated with 10 μm 8-bromo-cAMP (Sigma) or various concentrations of 7α-hydroxycholesterol (7α-OHC) (Steroloids, Inc., New Port, RI). Hemin (Sigma) solution was prepared in alkaline pH and diluted in 5% BSA solution. Cells were treated with 5 μm hemin-BSA solution. Luciferase reporters and expression plasmids were transfected into HepG2 cells using Tfx-20 or TransFast Reagent (Promega) following the manufacturer's instructions. Luciferase reporter assays were performed as previously described (30). Assays were performed in triplicate, and each experiment was repeated at least three times. Data were expressed as the means ± S.E.
Immunoblot Analysis
Total tissue and cell lysates were prepared in RIPA buffer (Cell Signaling Biotec, Inc., Boston, MA), and proteins were resolved on 10% SDS-PAGE gels. Monoclonal antibodies to human RORα (BioLegend, Inc., San Diego, CA), PKA, phosphorylated-PKA (P-PKA) (Cell Signaling Biotechnology Inc.), and phosphorylated serine (P-S) (Santa Cruz Biotechnology Inc., Santa Cruz, CA, and Millipore, Billerica, MA) were used for immunoblot analysis. For analysis of the half-life of RORα protein, RORα band intensity in immunoblots was determined as described previously (32). Half-life was calculated using the formula, half-life = ln2/−K, where K is the slope of the graph.
Immunoprecipitation Assay
HepG2 cells were transfected with the catalytic domain of a PKA-expressing plasmid for 24 h or treated with 8-bromo cAMP (10 μm) for 6 h with or without transfection of PKA-expressing plasmid. Immunoprecipitation assays were performed as described previously using RORα (BioLegend Inc.) antibody (30). Phosphorylation of serine in RORα was detected with phosphoserine (Ser(P))-specific antibody (Cell Signaling Technology, Inc.).
siRNA Transfection
RORα siRNA and scrambled control siRNA were purchased (Dharmacon Scientific) and transfected using Trans-fact Reagent (Qiagen Biolabs) for 4 h according to the manufacturer's protocol. Expression of RORα protein in liver lysates was analyzed using immunoblot.
Mammalian Two-hybrid Assay
For mammalian two-hybrid assays, the luciferase reporter plasmid p5xUAS-TK-Luc containing five copies of the upstream activating sequence (UAS) with the GAL4 binding site fused to the upstream of the thymidine kinase (TK) promoter was co-transfected with VP16-RORα and Gal4 fusion plasmids, Gal4-PGC1α, Gal4-CBP, or Gal4-SRC1 into HepG2 cells. Reporter activity was assayed as described previously (31).
Chromatin Immunoprecipitation Assay (ChIP)
Nuclei were isolated from mouse liver, and ChIP assays were performed with a kit (Millipore) as described previously (30). Antibodies against RORα (BioLegend, Inc.), acetyl-histone 3 (Cell Signaling Technology), and CBP (Santa Cruz Biotechnology) were used to immunoprecipitate chromatin. SYBR Green primers for real time PCR analysis of mouse CYP8B1 promoter were: −600TCCTCCCTGTGCCAGCTAAC (forward) and −416AAGGTTCCTGCCCTTGGACTT (reverse).
Bile Acid Analysis
Bile acids in liver, intestine (with content), and gallbladder were extracted with 95% ethanol overnight, 80% ethanol for 2 h, and methanol/chloroform (2:1) for 2 h at 50 °C. Total bile acids were determined with a bile acid assay kit (Genzyme Diagnostic, Framingham, MA). The bile acid pool was determined as the total amount of bile acids in liver, intestine, and gallbladder. Bile acid compositions in the gallbladder bile were analyzed using LC/MS/MS reported previously (33).
Statistical Analysis
Results are expressed as the mean ± S.E. except where indicated. Statistical analysis was performed by Student's t test. p < 0.05 was considered statistically significant.
RESULTS
Fasting Induces CYP8B1 and RORα Expression in Hepatocytes
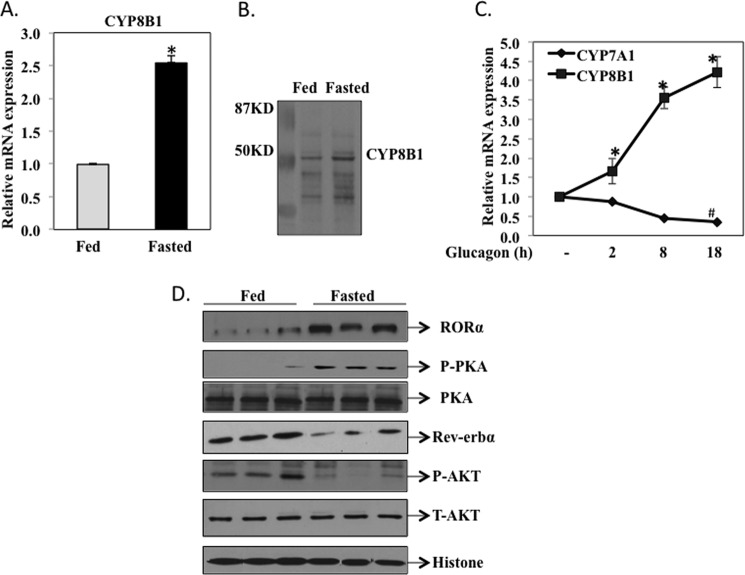
To study fasting-induced changes in gene expression involved in bile acid synthesis, wild type mice (C57Bl/6J) were fasted for 12 h (ZT2 to ZT14) and then fed normal chow for 6 h and sacrificed at ZT20. A quantitative real time PCR assay (Fig. 1A) shows that CYP8B1 mRNA was 2.5-fold higher in fasted mice compared with fed mice. Immunoblot analysis shows that CYP8B1 protein expression levels were higher in the livers of fasted than fed mice (Fig. 1B). We then assayed the effect of the fasting hormone glucagon on CYP8B1 mRNA expression in primary human hepatocytes. Fig. 1C shows that CYP8B1 mRNA levels were rapidly increased in human primary hepatocytes treated with glucagon (100 ng/ml), whereas CYP7A1 mRNA levels were reduced as expected. Because RORα is a fasting-induced gene, we investigated whether there was a correlation between CYP8B1 and RORα expression levels in mouse liver. Western immunoblot analysis (Fig. 1D) shows that RORα protein levels were significantly higher in fasted than fed mice, which is opposite that of Rev-erbα, a negative core clock gene. Glucagon/cAMP signaling is known to activate PKA by phosphorylation during fasting. Fig. 1D shows that phosphorylation of PKA was increased in fasted mice as expected, whereas phosphorylation of AKT/PKB in insulin signaling was induced in fed mice. These data suggested that fasting and glucagon signaling induced CYP8B1 and RORα protein levels but reduced Rev-erbα protein levels in mouse liver.
FIGURE 1.
Fasting induces CYP8B1 mRNA and RORα protein expression in hepatocytes. Male C57BL6J mice (12 weeks old) were fasted for 12 h from 8 a.m. (ZT2) to 8 p.m. (ZT14). A group of 4 mice was sacrificed at ZT14 as the fasted group, and another group (4) was re-fed a regular chow diet for 6 h and sacrificed at 2 am (ZT20) as the fed group. A, quantitative real-time PCR analysis of CYP8B1 mRNA expression in mouse liver. Relative CYP8B1 mRNA expression in fasted mouse liver was calculated with respect to fed mice. Results are expressed as the mean ± S.E.; n = 4; an asterisk indicates statistical significance, p < 0.05, fasted versus fed mice. B, Western immunoblot analysis of CYP8B1 protein in liver microsomes isolated from fasted and fed mice (pooled from n = 4). C, real-time PCR analysis of CYP8B1 and CYP7A1 mRNA expression in human primary hepatocytes (donors #1839, #1958, and #1959) treated with glucagon (100 ng/ml). mRNA samples were isolated from cells at the time points indicated. Results were expressed as mean ± S.E.; n = 3; * and # indicate statistical significance, p < 0.05, glucagon treated versus vehicle-treated cells. D, Western immunoblot analysis of RORα, Rev-erbα, PKA, p-PKA, AKT, and p-AKT in total liver extracts of fasted (16 h) and fed (6 h) mice. Each lane was loaded with liver extracts from different mice in each group. Histone 3 was used as a loading control.
RORα Is Stabilized by PKA-mediated Phosphorylation
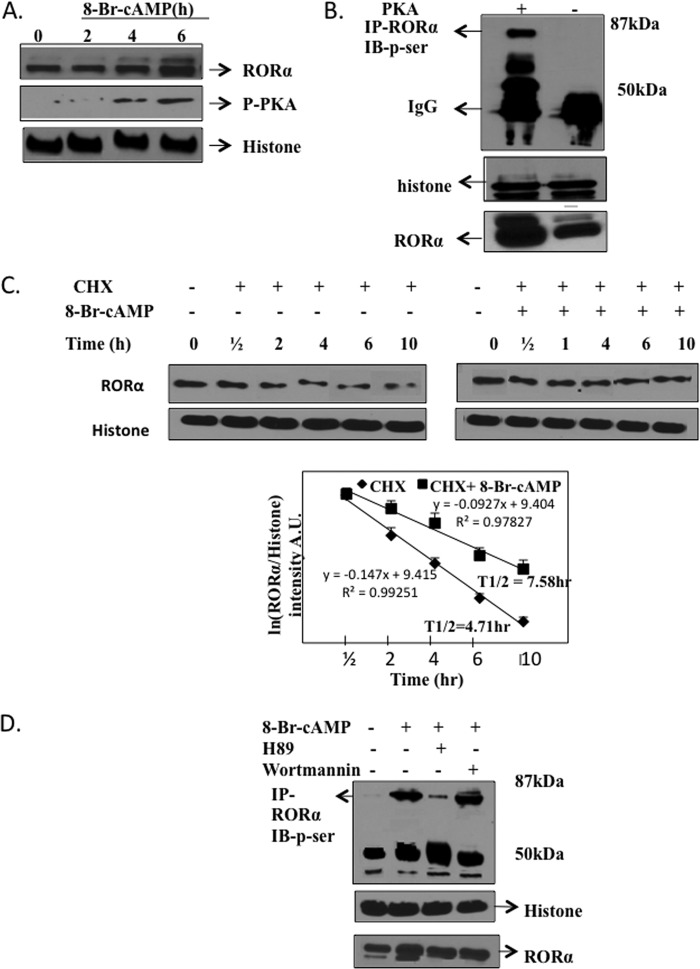
We hypothesize that the fasting signaling pathway glucagon/cAMP/PKA may phosphorylate and stabilize RORα protein. To test the effect of cAMP on RORα protein expression, we treated HepG2 cells with 8-Br-cAMP and monitored RORα protein expression by Western blot analysis. Results in Fig. 2A clearly showed that 8-Br-cAMP treatment increased RORα protein and phosphorylated PKA. We then performed an in vitro phosphorylation assay of RORα using HepG2 cells transfected with a catalytically active PKA expression plasmid and an RORα antibody to immunoprecipitate RORα. Then, an antibody against P-Ser was used to detect phosphorylation of RORα. Fig. 2B showed that RORα was phosphorylated by PKA. To further confirm that cAMP increased RORα protein stability, we treated HepG2 cells with cycloheximide (a protein synthesis inhibitor) with or without treatment of 8-Br-cAMP and monitored RORα protein levels at different time points. Fig. 2C shows that RORα protein levels were diminished with cycloheximide (CHX) treatment in a time-dependent manner (left panel), and treatment with 8-Br-cAMP significantly prevented reduction of RORα protein levels (right panel). The calculated half-life of RORα was 4.71 h and 8-Br-cAMP increased RORα half-life to 7.58 h (Fig. 2C, lower figure). These experiments provided evidence that the glucagon/cAMP/PKA pathway phosphorylated RORα and increased its half-life to induce CYP8B1 in fasting response. We further studied the specificity of cAMP/PKA phosphorylation of RORα. Immunoprecipitation assays (Fig. 2D) show that the PKA inhibitor H89 blocked cAMP-mediated phosphorylation of RORα, but the phosphoinositol 3-kinase inhibitor wortmannin had no effect.
FIGURE 2.
Phosphorylation of RORα by PKA stabilizes RORα protein. A, Western blot analysis of HepG2 cell extracts (total) treated with 8-Br-cAMP (10 μm) or vehicle (DMSO) using an antibody against RORα, phosphorylated-PKA (p-PKA), or histone-3 (control). B, immunoprecipitation (IP) assay of phosphorylation of RORα by PKA. HepG2 cells were transfected with a catalytically active PKA expression plasmid (0.1 μg) or control plasmid. Cell extracts were incubated with an antibody against RORα to precipitate RORα, and precipitates were subject to SDS-gel electrophoresis. Phosphorylation of RORα was detected using an antibody against p-Ser. IB, immunoblot. C, analysis of the half-life of RORα. HepG2 cells were treated with cycloheximide (CHX, 20 μm) with or without 8-Br-cAMP (10 μm) for the times indicated. Cell extracts were subject to SDS gel electrophoresis and immunoblotted with an antibody against RORα or histone 3 (control). The bottom panel shows a first order reaction plot of ln(RORα/histone band intensity) versus time (h). Slope (−K) of the line was calculated with an equation, t½ = ln 2/−K. A.U., absorbance units. D, immunoprecipitation assay of specificity of cAMP/PKA-stimulated phosphorylation of RORα. HepG2 cells were treated with 8-Br-cAMP and a PKA inhibitor H89 or a phosphoinositol 3-kinase inhibitor, wortmannin, at 10 μm, as indicated. Cell lysates were immunoprecipitated with an antibody against RORα and subjected to SDS-gel electrophoresis and blotted with a specific antibody against Ser(P). Immunoblots of histone and RORα were used as loading controls.
Diurnal Expression of CYP8B1 and RORα in Mouse Liver
RORα is a core clock gene that is known to regulate liver metabolism and circadian rhythm of many metabolic genes in the liver (34). We thus studied the Diurnal rhythm of CYP8B1 and RORα expression and the effect of restricted feeding. In free-fed mice (Fig. 3A), CYP8B1 mRNA levels showed a diurnal rhythm, decreased during the day and reaching the lowest levels 2 h into the dark cycle (ZT14). This circadian rhythm pattern is exactly opposite that of CYP7A1, which shows a peak after the onset of the dark cycle (15). During restricted feeding, mice were fasted from ZT2 to ZT18, allowed to eat for 3 h, and sacrificed at ZT21. Fasting gradually induced CYP8B1 mRNA levels (upper panel, Fig. 3) and restricted feeding at ZT18 rapidly reduced CYP8B1 mRNA levels, thus altering the diurnal rhythm of CYP8B1. We studied the rhythm of mRNA expression of RORα and Rev-erbα. These two clock genes had different rhythms in expression; the positive clock gene RORα peaked at ZT14 (middle panel, Fig. 3), whereas the negative clock gene Rev-erbα peaked at ZT6 (mid-day) (lower panel, Fig. 3). Thus, the increase of RORα coincided with increased CYP8B1 mRNA in fasting, which is opposite the rhythm of Rev-erbα mRNA. It is noted that fasting and restricted feeding did not alter the phase of expression of these two clock genes, only the amplitude.
FIGURE 3.
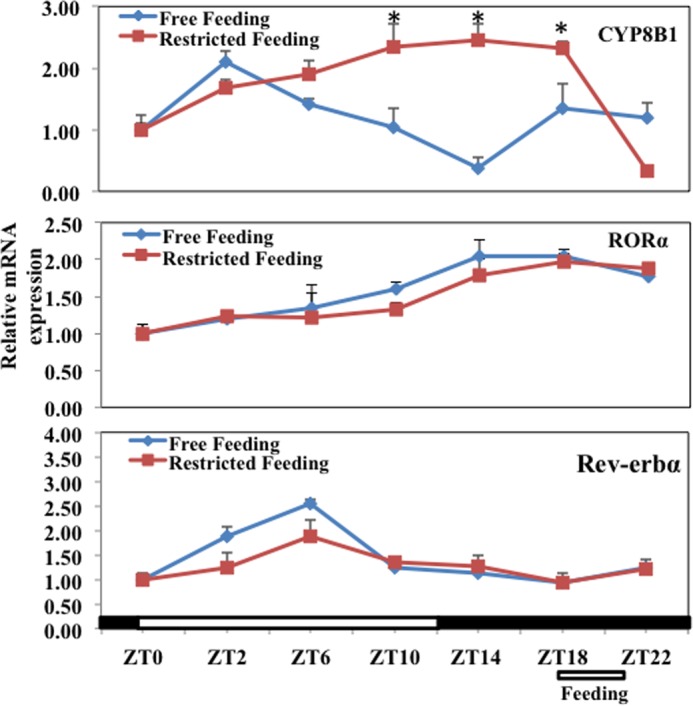
Effect of fasting and restricted feeding on diurnal rhythm of CYP8B1, RORα, and Rev-erBα mRNA expression in mouse liver. Diurnal rhythm of liver CYP8B1 mRNA expression (upper panel), RORα mRNA expression (middle panel), and Rev-Erbα mRNA expression in mouse liver (lower panel). Male C57BL6J mice (12–18 weeks old) either had free access to food for 24 h (Free feeding) or were fasted at ZT2 and then had restricted access to food at ZT18 for 3 h (Restricted feeding) as indicated. Mice were sacrificed every 4 h (n = 4/5 at each time point). Lights were on at ZT0 and off at ZT12. Relative mRNA expression was calculated with respect to ZT0. Results were expressed as the mean ± S.E.; n = 4; asterisks indicate statistical significance, p < 0.05, fasted-fed versus free-fed mice.
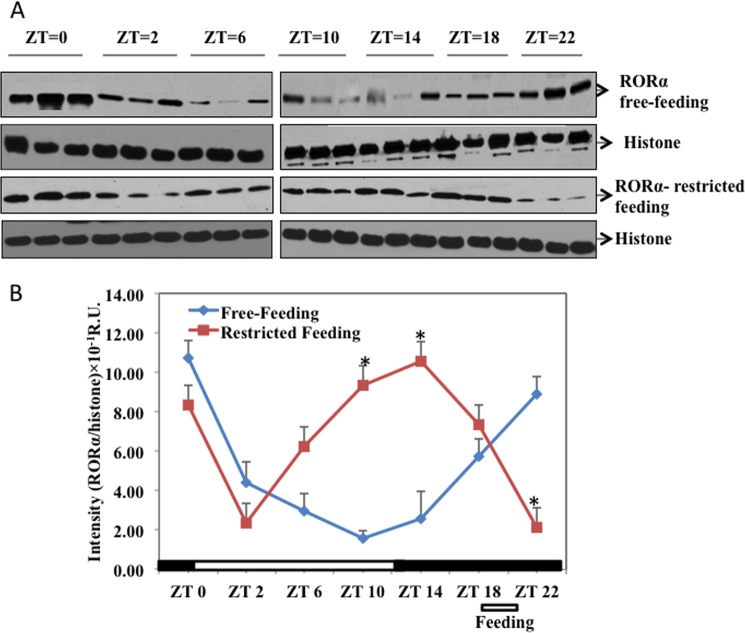
Immunoblot analysis (Fig. 4) shows that in free feeding, RORα protein levels decreased to the lowest level at ZT10 and then gradually increased and peaked at ZT22, about 8 h after the peak of RORα mRNA. However, in restricted feeding, fasting gradually increased RORα protein levels and reached the highest levels at ZT14. Feeding at ZT18 rapidly decreased RORα protein levels. This pattern of RORα protein expression is different from its mRNA expression, suggesting post-transcriptional regulation of RORα. This is consistent with stabilization of RORα protein by phosphorylation (Fig. 2). The changes of RORα protein levels (Fig. 4B) correlated well with changes of CYP8B1 mRNA levels (top panel, Fig. 3), suggesting that RORα regulates CYP8B1 mRNA transcription.
FIGURE 4.
Restricted feeding changes RORα protein expression in mouse liver. A, total liver lysates were isolated from mice sacrificed at time points indicated. Samples (three at each time point) were loaded onto two separate gels run at the same time. B, intensity (absorbance units (A.U.)) of the RORα protein band (average of three samples) was normalized to histone as the loading control at each time point. Asterisks indicate statistical significance, p < 0.05, restricted feeding versus free-fed mice. R.U., relative units.
RORα Stimulated CYP8B1 mRNA Levels in Mouse Liver and Increased 12α-Hydroxylated Bile Acids in Bile
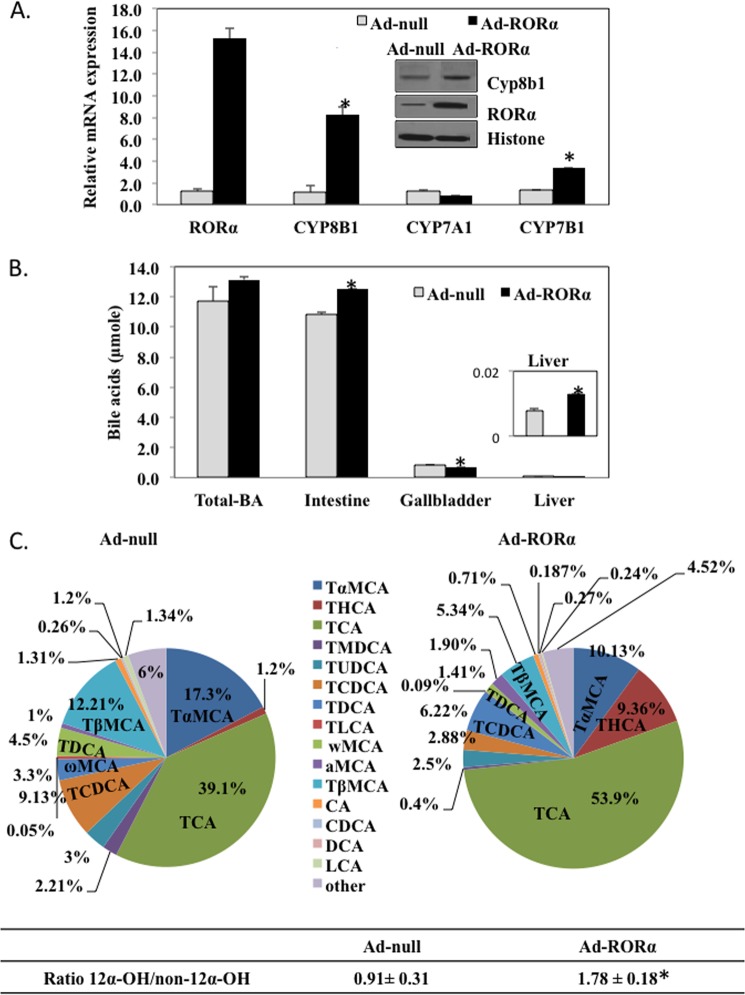
To study whether RORα stimulates CYP8B1 gene expression in vivo, we generated an adenovirus expressing human RORα (Ad-RORα) to administer RORα to mouse liver and assayed CYP8B1 expression levels. Fig. 5A showed that Ad-RORα markedly increased CYP8B1 and CYP7B1 mRNA by 8- and 3-fold, respectively. Ad-RORα did not change CYP7A1 mRNA. RORα overexpression induced RORα and CYP8B1 protein levels in mouse liver microsomes (inset in Fig. 5A).
FIGURE 5.
Adenovirus-mediated expression of RORα induces CYP8B1 mRNA and protein levels in mouse liver. A, effect of adenovirus-mediated transduction of RORα on CYP8B1 mRNA expression in mouse liver. Mice (C57Bl/6J, n = 6) were injected (intravenously) with adenovirus expressing RORα (Ad-RORα) (2 × 109 pfu/mouse) or same amount of empty adenovirus (Ad-null). Relative mRNA expression was calculated with respect to Ad-null injected mice. Inset, Western immunoblot analysis of RORα and CYP8B1 protein in microsomes isolated from livers pooled from Ad-RORα infected mice (n = 6) and control (Ad-null n = 6) mice. B, analysis of total bile acid pool size including liver, gallbladder, and intestine of Ad-RORα and Ad-null infected mice. Results were expressed as the mean ± S.E.; n = 6; asterisks indicate statistical significance, p < 0.05, Ad- RORα versus Ad-null infused mice. C, gallbladder bile acid analysis. Wild type C57BL6 mice were injected with Ad-null (n = 3) or Ad-RORα (n = 6). Seven days post injection, gallbladder bile was extracted for analysis of bile acid composition using LC/MS/MS as described under “Experimental Procedures.” Each bile acid was calculated as % of total bile acids measured (22284.65 pm). The ratio of 12α-hydroxylated bile acid to non-12α-hydroxylated bile acids was calculated. 12α-Hydroxylated bile acid species are CA, deoxycholic acid, tauro-cholic acid (TCA), and tauro-deoxycholic acids (TDCA). Non-12α-hydroxylated bile acid species are tauro-chenodeoxycholic acid (TCDCA), TαMCA, TβMCA, tauro-urodeoxycholic acids (TUDCA), tauro-hyocholic acid (THCA), tauro-murideoxycholic acid (TMDCA), tauro-lithocholic acid (TLCA). The asterisk indicates statistical significance, p < 0.05, Ad-RORα versus Ad-null infused mice.
Ad-RORα did not alter the total bile acid pool in the enterohepatic circulation (Fig. 5B) but significantly increased bile acid content in the intestine and liver and significantly decreased that in gallbladder bile. We analyzed bile acid compositions in gallbladder bile. Tauro-cholic acid content was increased from 39.1% in Ad-null to 54% in Ad-RORα mice. Tauro-chenodeoxycholic acid was reduced from 9.1% in Ad-null to 2.9% in Ad-RORα mice. In mouse liver, CDCA (3α, 7α) is converted to highly hydrophilic bile acids, tauro-α-muricholic acid (TαMCA) (3α, 6β, and 7α), tauro-β-muricholic acid (TβMCA) (3α, 6β, and 7β), tauro-hyocholic acid (3α, 6α, and 7α), and tauro-ursodeoxycholic acid (3α, 7β) by 6-hydroxylation catalyzed by Cyp3a11 and/or by epimerization reactions. TαMCA and TβMCA were decreased from 17.3 and 12.2% in Ad-null mice to 10.1 and 5.3% in Ad-RORα mice, respectively. It is interesting that tauro-hyocholic acid was markedly increased from 1.2% in Ad-null to 9.4% in Ad-RORα mice. The ratio of total 12α-hydroxylated bile acids (tauro-cholic acid, tauro-deoxycholic acids, CA, and deoxycholic acid) to non-12α-hydroxylated bile acids was increased from 0.91 in Ad-null mice to 1.78 in Ad- RORα mice, as the result of increased CYP8B1 activity in Ad-RORα mice. We analyzed serum and liver cholesterol and triglyceride levels upon RORα overexpression. Ad-RORα significantly increased serum cholesterol contents by ∼30% (Fig. 6A) and showed a tendency to increase liver cholesterol content (Fig. 6B) but had no effect on serum (Fig. 6C) and liver triglyceride contents (Fig. 6D) in mice.
FIGURE 6.
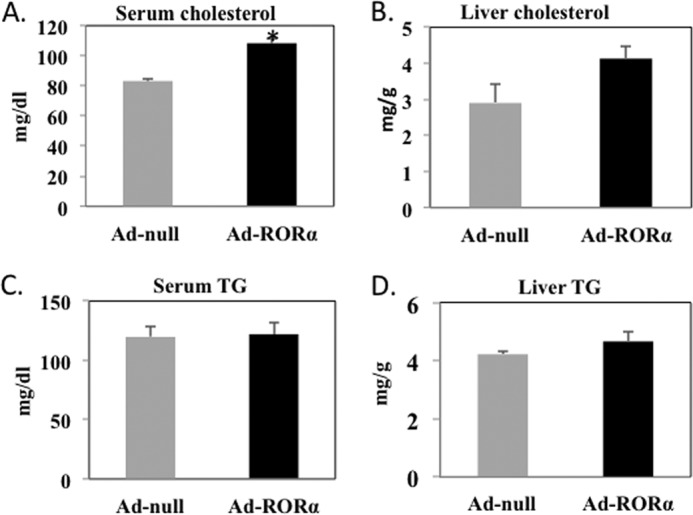
Analysis of lipid profile of mice transfected with Ad-RORα. A, serum cholesterol. B, liver cholesterol. C, serum triglyceride. D, liver triglyceride. Wild type C57BL6 mice (n = 5 to 6) were injected with Ad-RORα or Ad-null as described under “Experimental Procedures.” Seven days post injection mice were fasted overnight. Fresh serum and livers were obtained for analysis of triglyceride (TG) and cholesterol as described under “Experimental Procedures.” Results are expressed as the mean ± S.E.; n = 5; the asterisk indicates statistical significance, p < 0.05, Ad-RORα versus Ad-null infected mice.
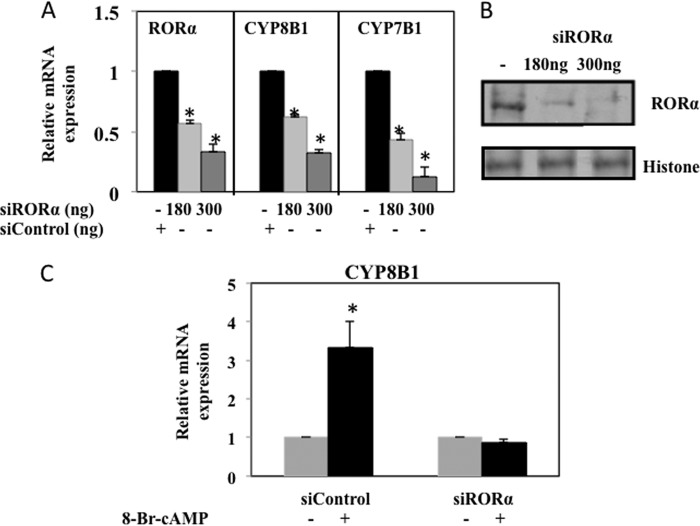
Furthermore, we used siRNA to knock down RORα expression in HepG2 cells and assayed CYP8B1 mRNA expression levels. RORα-siRNA efficiently knocked down RORα mRNA (Fig. 7A) and protein (Fig. 7B) expression in HepG2 cells. RORα-siRNA also efficiently knocked down CYP8B1 and CYP7B1 mRNA levels by ∼70% compared with cells transfected with scrambled siRNA (Fig. 7A). RORα-siRNA also prevented 8-Br-cAMP induction of CYP8B1 mRNA (Fig. 7C). These data suggest that fasting and cAMP induction of CYP8B1 is RORα-dependent.
FIGURE 7.
siRNA-mediated knockdown of RORα decreased CYP8B1 mRNA expression. A, HepG2 cells were transfected with RORα siRNA (180 and 300 ng) for 8 h to knockdown RORα expression. Scrambled siRNA to RORα (siControl 300 ng) was used as a negative control. Relative mRNA expression of CYP8B1, CYP7B1, and RORα was assayed by real-time PCR. B, Western blot analysis of RORα protein (10% acrylamide gel) using total HepG2 cell lysate transfected with RORα siRNA (180 and 300 ng) for 10 h. Scrambled siRNA (300 ng) (siControl) was used as a negative control. C, HepG2 cells were transfected with siRORα (180 and 300 ng) or scrambled siControl as a negative control for 4 h followed by treatment of 8-Br-cAMP (10 μm) for 8 h. Scrambled siControl was used as a negative control. Real-time PCR was used to assay relative mRNA expression of CYP8B1. Results were expressed as the mean ± S.D.; n = 3; the asterisk indicates statistical significance, p < 0.05, siControl versus siRORα transfected cells.
Identification of a Functional RORα Binding Site in the CYP8B1 Gene Promoter
We wanted to elucidate the molecular mechanism of regulation of CYP8B1 gene expression by RORα. A transcription factor sequence motif search identified a putative RORE (+22AAAAGGTCA+30) located in the 5′ upstream region of human CYP8B1 gene promoter. A human CYP8B1 (−514/+300) promoter-Luciferase reporter plasmid containing this RORE was co-transfected with a RORα expression plasmid. Fig. 8A showed that RORα stimulated human CYP8B1-Luc reporter activity by 6-fold, and 7α-OHC antagonized the stimulatory effect of RORα by about 20% at 10 μm. On the other hand, Rev-erbα significantly reduced CYP8B1-Luc reporter activity in the absence or presence of heme, which is an endogenous ligand of Rev-erbα. Fig. 8C showed 7α-OHC dose-dependently inhibited CYP8B1 mRNA expression levels, whereas 25-hydroxycholesterol had no effect on CYP8B1 mRNA levels in HepG2 cells. To confirm the function of this putative RORE, we mutagenized the RORE sequence in the reporter plasmid and showed that mutant CYP8B1-Luc reporter did not respond to induction by RORα (Fig. 8D).
FIGURE 8.
Identification of a RORα response element in human CYP8B1 gene promoter. Transient transfection assay of human CYP8B1-Luc reporter activity. HepG2 cells were co-transfected with a human CYP8B1 (−514/+300)-Luc reporter (0.2 μg) and a RORα (0.1 μg) (A) and Rev-erbα (0.1 μg) expression plasmid or a pGL3 basic empty plasmid (B). Cells were treated either with 7α-hydroxycholesterol (20 μm), hemin (5 μm), or DMSO and BSA (5%) as vehicle control. Luciferase activity was measured, and luciferase activities (RLU) were normalized to β-galactosidase (β-gal). Each transfection was performed in triplicate. Results were expressed as the mean ± S.D.; n = 3; asterisks indicate statistical significance, p < 0.05, 7α-OHC-treated versus vehicle (DMSO)-treated control cells and hemin-treated versus vehicle (5% BSA). C, effect of 7α-hydroxycholesterol on CYP8B1 mRNA expression in HepG2 cells. Cells were treated with various concentrations 7α-OHC or 25-hydroxycholesterol (25-OHC) as indicated. Real-time PCR was used to assay relative mRNA expression of CYP8B1 in cells treated with 7α-OHC, 25-hydroxycholesterol, or vehicle (DMSO) at the concentrations indicated. Results were expressed as the mean ± S.E., asterisks indicate statistical significance, p < 0.05, treated versus vehicle (DMSO) control. D, mutagenesis analysis of a putative RORα response element (RORE) in human CYP8B1 gene promoter. Diagrams on the top show DNA sequences of wild type and mutant RORE in human CYP8B1 promoter-Luc reporters. Cells were transfected with wild type or mutant CYP8B1 reporter plasmid (0.2 μg) with or without RORα plasmid (0.1 μg). Each transfection was performed in triplicate. Results are expressed as the mean ± S.D., an asterisk indicates statistical significance, p < 0.05, RORα co-transfected versus empty plasmid transfected cells. E, EMSA of RORα binding to RORα response element on CYP8B1 promoter. 32P-Labeled probes (specific activity = 80,000 cpm) were mixed with in vitro transcription and translation lysates programmed with a human RORα expression plasmid in binding buffer (80 mm HEPES, 0.8 mm EDTA, 40% glycerol, 16 mm MgCl2 with 10 mm DTT and 1 μl of dI-dC added just before the reaction). A/T-rich region mutant (M1) and RORE half-site mutant (M2) were tested for RORα binding. For cold completion, non-radiolabeled APOCIII probe (100×) was added. For antibody super shift assay, a RORα antibody (3 μl) was added in binding buffer without DTT (sixth lane) in EMSA assay mixture. Samples were loaded on a non-denaturing gel (10% polyacrylamide) for electrophoresis.
To verify if RORα binds to this putative RORE in the human CYP8B1 gene promoter, we performed EMSA. Fig. 8E showed that using a labeled probe design based on this RORE, in vitro translated RORα formed a complex with the probe with similar mobility as the RORE probe designed according to a canonical RORE in the APOCIII gene. Mutations of the RORE sequences in the human CYP8B1 gene (M1, mutated the core AGGTCA sequence; M2, mutated the 5′ A/T-rich sequence) abolished the binding of RORα. Competition assays showed that a 3× excess of unlabeled APOCIII probe abolished mobility shift. The addition of an antibody against RORα retarded the mobility of the DNA-RORα complex (Fig. 8E). We also performed similar experiments and identified a conserved RORE motif in mouse CYP8B1 gene promoter (data not shown). These data suggest that the RORα binding site on the human CYP8B1 gene promoter is functional, and RORα binds to this RORE and stimulates human CYP8B1 gene transcription.
RORα Interacts with CBP to Stimulate cyp8b1 Gene Transcription
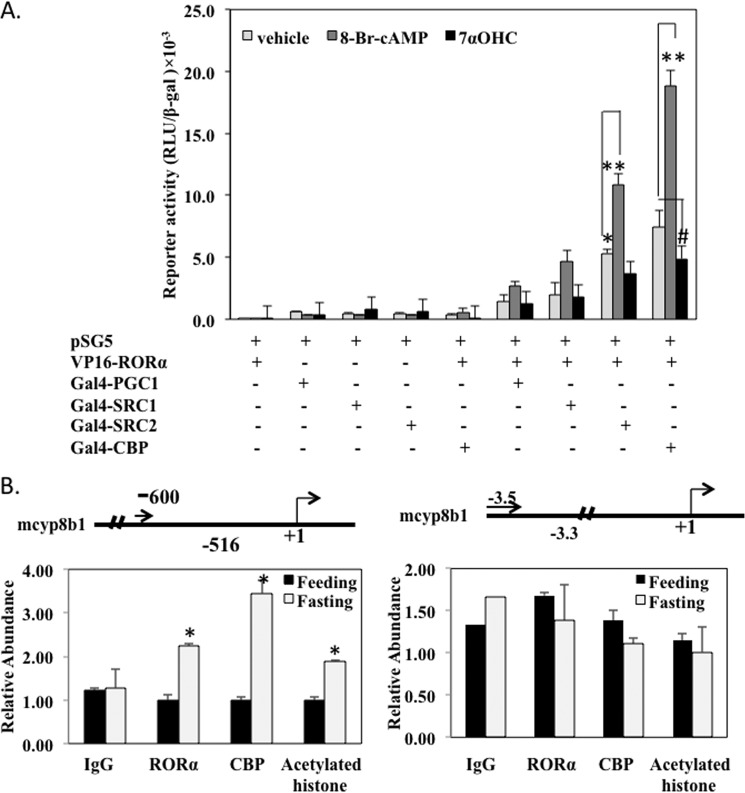
To study trans-activation of the CYP8B1 gene by RORα and co-activators, we performed a mammalian two-hybrid assay to study the interaction of RORα with several nuclear receptor co-activators. In this assay a GAL4 reporter pSG5 containing five copies of the upstream activation sequence of the Gal4 binding site fused upstream of the thymidine kinase gene promoter, and the luciferase gene was co-transfected with a Gal4 fusion protein plasmid and VP16-RORα plasmid. If the Gal4 fusion protein binds to the reporter, it interacts with VP16 fusion to stimulate Gal4 reporter activity. Fig. 9A showed that the interaction of RORα with CBP was the strongest among five co-activators tested. Furthermore, 8-Br-cAMP strongly increased, whereas 7α-OHC decreased RORα interaction with co-activators. Overall, these experiments suggest that CBP is the most efficacious co-activator that interacts with RORα to stimulate CYP8B1 gene transcription, and cAMP stimulated CBP interaction with RORα, whereas 7α-OHC antagonized RORα activity to inhibit its interaction with CBP.
FIGURE 9.
RORα interaction with CBP and stimulates histone acetylation of CYP8B1 promoter. A, mammalian two-hybrid assay of RORα interaction with co-activators. A GAL4-TK-Luc reporter, pSG5 (0.2 μg) was co-transfected with VP16-RORα (0.1 μg) and 0.1 μg of Gal4-PGC1α, Gal4-SRC1, Gal4-SRC2, or Gal4-CBP fusion plasmid in HepG2 cells. Cells were treated with 7α-OHC (20 μm) or 8-bromo-cAMP or vehicle for 12 h. Luciferase activity was measured and expressed as relative light units (RLU) normalized to β-galactosidase activity. Each transfection was performed in triplicate. Results are expressed as the mean ± S.E., asterisks indicate statistical significance, p < 0.05, 8-Br-cAMP- or 7α-OHC-treated versus vehicle (DMSO) cells. B, chromatin immunoprecipitation assay of RORα and CBP on Cyp8b1 promoter. Mouse livers from the same experiments under Fig. 1 were used to isolate nuclei for ChIP assay. For precipitation of chromatin, nuclei were mixed with an antibody (8 μl) against RORα, CBP, mono-acetylated histone, or hepatocyte nuclear factor 4α (HNF4α), and rabbit IgG were used as negative control. Left panel, chromatin occupancy (Relative Abundance) was analyzed using real-time PCR to amplify a region of mouse Cyp8b1 promoter containing the RORE (shown in the upper panel). The right panel is the negative control of a 5′-upstream region of mouse Cyp8b1 promoter. Data were normalized using 10% input, and relative chromatin occupancy was calculated with respect to control adenovirus treatment (Ad-null). Results were expressed as the mean ± S.E.; n = 4 for each set of adenovirus over expression; asterisks indicate statistical significance, p < 0.05, Ad-RORα- versus Ad-null-infected mice.
We then used a ChIP assay to study the binding of RORα and CBP to the CYP8B1 gene promoter region containing the putative RORα binding site. We fasted mice for 12 h (ZT14) and fed them for 6 h (ZT20). Liver nuclear extracts were isolated for ChIP assay using a specific antibody against RORα or CBP to precipitate RORα-DNA or RORα/CBP-DNA complexes. Primers were designed to amplify a fragment from nucleotide −600 to the −516 region containing the RORE. Fig. 9B, left panel, showed that RORα binding to the CYP8B1 promoter was significantly increased when nuclear extract isolated from fasted mice liver was compared with liver from fed mice. CBP is a co-activator that does not bind DNA; thus, CBP was recruited by RORα to the CYP8B1 promoter. Because CBP is a histone acetylase, it increased acetylation of histone on the CYP8B1 gene promoter (Fig. 9B). A region 5′ upstream was assayed as a negative control for ChIP assay (Fig. 9B, right panel).
DISCUSSION
In this study we investigated the underlying molecular mechanism of diurnal and fasting regulation of CYP8B1 gene expression. CYP8B1 expression exhibits a diurnal rhythm that is opposite that of CYP7A1 and bile acid synthesis. Feeding rapidly and strongly inhibited CYP8B1 mRNA and protein expression but had no effect on rhythmic RORα mRNA expression. Interestingly, feeding strongly reduced RORα protein expression levels, which correlated to reduction of CYP8B1 expression. Thus, regulation of RORα protein stability plays a key role in diurnal and fasting-refeeding responses of CYP8B1. Fasting induces CYP8B1 expression in mouse liver via glucagon/cAMP/PKA signaling, which phosphorylates and increases RORα half-life. RORα also increases histone acetylation of the CYP8B1 promoter by recruiting a histone acetylase, CBP. In contrast, Rev-erbα is a negative clock gene that recruits a nuclear receptor co-repressor 1 (NCoR1) and histone deacetylase 3 to inhibit target gene expression by binding to the same response element as RORα (35). Thus, Rev-erbα and RORα oscillate to regulate the circadian rhythm of CYP8B1 and also in feeding and fasting response of CYP8B1.
ROR-mediated circadian and nutrient regulation of CYP8B1 may play an important role in maintaining overall bile acid composition. Increasing evidence suggests that not only the total bile acid pool size but also the bile acid composition play critical roles in regulating metabolic homeostasis, and altered bile acid composition has been implicated in the pathogenesis of metabolic syndromes (27). The ratio of 12α-hydroxylated to non-12α-hydroxylated bile acids in bile is important in intestinal absorption of dietary cholesterol. Although increased CA and bile acid pool have been shown in diabetic patients (36) and in diabetic mouse models (27), the underlying mechanism is still not clear. This study shows that RORα may be a key activator of CYP8B1 to increase 12-hydroxylated bile acids and contribute to increased serum and liver cholesterol contents. Overexpression of CYP8B1 may decrease hepatic production of CDCA and may also decrease FXR and TGR5 signaling, resulting in dyslipidemia, obesity, and insulin resistance. In contrast, a recent study of liver-specific forkhead transcription factor O1 (FoxO1) knock-out mice in an LDL receptor knock-out background shows reduced 12α-hydroxylated bile acids and increased hepatic steatosis and hepatic insulin resistance, suggesting that deficiency of 12α-hydroxylated bile acids and CYP8B1 in these mice hinders the triglyceride-lowering effects of FXR (37). FoxO1 is known to mediate insulin signaling in regulation of gluconeogenesis and other target genes (38, 39). FoxO1 also can act as a co-repressor of PGC-1α and hepatocyte nuclear factor 4α (HNF4α), thus inhibiting transcription of PGC-1α and HNF4α-regulated genes, including CYP7A1 and CYP8B1 (40, 41). Insulin is known to inhibit CYP8B1 (11). Thus, insulin action via FoxO1 should inhibit, not induce, CYP8B1 as suggested from a liver-specific FoxO1 knock-out mouse study (37). Further study of the role of FoxO1 in regulation of bile acid synthesis is needed to understand the role of CYP8B1 in hepatic steatosis.
This RORα-mediated CYP8B1-overexpressing mouse model could be useful for studying the pathogenesis of Non-alcoholic fatty liver disease (NAFLD), atherosclerosis, and diabetes. This mouse model is opposite to the CYP7A1 transgenic mouse model, which is resistant to Western high fat diet-induced insulin resistance and obesity (42). In CYP7A1 transgenic mice, bile acid pool size increased 2-fold, and CYP8B1 was inhibited and resulted in CDCA as the predominant bile acid with very little CA in the pool. Studies of these two mouse models of bile acid synthesis provide strong evidence that both bile acid pool size and composition are important in regulation of hepatic lipid metabolism and metabolic diseases. Hydrophobic bile acids are toxic and can cause inflammation and injury in the liver and intestine. The balance of bile acid pool size with proper bile acid composition may be important in activating FXR and TGR5 signaling for anti-inflammation in the digestive system. On the other hand, enlarging the bile acid pool with CA may cause dyslipidemia and contribute to cholesterol gallstone disease and non-alcoholic fatty liver disease, which are highly prevalent in type II diabetic patients.
It is interesting that 7α-OHC could act as an inverse agonist of RORα and suppress CYP8B1 gene transcription. It is possible that increasing CYP7A1 activity may antagonize RORα induction of CYP8B1 and result in altered bile acid composition and rate of bile acid synthesis in circadian rhythm (15) and the fasting-refeeding response of these two regulatory genes in bile acid synthesis. On the other hand, activation of RORα by cholesterol may be a mechanism for promoting hepatic inflammation and steatosis and insulin resistance by dietary cholesterol (19, 21). RORα plays a key role in integrating the circadian clock to liver metabolism, immunity, and inflammation (34). This study contributes to our understanding of the molecular mechanism by which bile acid synthesis and composition regulates hepatic metabolic homeostasis. Antagonizing RORα activity may be a potential therapeutic strategy for treating inflammatory diseases such as non-alcoholic fatty liver disease, diabetes, and obesity.
Acknowledgment
We gratefully acknowledge Dr. Curt Klaassen of University of Kansas Medical School for analysis of bile acid composition.
This work was supported, in whole or in part, by National Institutes of Health Grants DK44442 and DK58379 (to J. Y. L. C.).
- CA
- cholic acid
- CDCA
- chenodeoxycholic acid
- CYP7A1
- cholesterol 7α-hydroxylase
- CYP7B1
- oxysterol 7α-hydroxylase
- CYP8B1
- sterol 12α-hydroxylase
- FoxO1
- forkhead transcription factor O1
- PGC1α
- peroxisome proliferator activated receptor γ co-activator 1α
- ROR
- retinoic acid-related orphan receptor α
- RORE
- ROR response element
- SRC
- steroid receptor co-activator
- CBP
- cAMP response element-binding protein-binding protein
- FXR
- farnesoid X receptor
- 7α-OHC
- 7α-hydroxycholesterol
- ZT
- Zeitgeber time
- TαMCA
- tauro-β-muricholic acids.
REFERENCES
- 1. Chiang J. Y. (2009) Bile acids. Regulation of synthesis. J. Lipid Res. 50, 1955–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Watanabe M., Houten S. M., Mataki C., Christoffolete M. A., Kim B. W., Sato H., Messaddeq N., Harney J. W., Ezaki O., Kodama T., Schoonjans K., Bianco A. C., Auwerx J. (2006) Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature 439, 484–489 [DOI] [PubMed] [Google Scholar]
- 3. Pols T. W., Noriega L. G., Nomura M., Auwerx J., Schoonjans K. (2011) The bile acid membrane receptor TGR5 as an emerging target in metabolism and inflammation. J. Hepatol. 54, 1263–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Thomas C., Pellicciari R., Pruzanski M., Auwerx J., Schoonjans K. (2008) Targeting bile-acid signalling for metabolic diseases. Nat. Rev. Drug Discov. 7, 678–693 [DOI] [PubMed] [Google Scholar]
- 5. Wang D. Q., Lammert F., Cohen D. E., Paigen B., Carey M. C. (1999) Cholic acid aids absorption, biliary secretion, and phase transitions of cholesterol in murine cholelithogenesis. Am. J. Physiol. 276, G751–G760 [DOI] [PubMed] [Google Scholar]
- 6. Murphy C., Parini P., Wang J., Björkhem I., Eggertsen G., Gåfvels M. (2005) Cholic acid as key regulator of cholesterol synthesis, intestinal absorption, and hepatic storage in mice. Biochim. Biophys. Acta 1735, 167–175 [DOI] [PubMed] [Google Scholar]
- 7. Li-Hawkins J., Gåfvels M., Olin M., Lund E. G., Andersson U., Schuster G., Björkhem I., Russell D. W., Eggertsen G. (2002) Cholic acid mediates negative feedback regulation of bile acid synthesis in mice. J. Clin. Invest. 110, 1191–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang J., Gåfvels M., Rudling M., Murphy C., Björkhem I., Einarsson C., Eggertsen G. (2006) Critical role of cholic acid for development of hypercholesterolemia and gallstones in diabetic mice. Biochem. Biophys. Res. Commun. 342, 1382–1388 [DOI] [PubMed] [Google Scholar]
- 9. Wang J., Einarsson C., Murphy C., Parini P., Björkhem I., Gåfvels M., Eggertsen G. (2006) Studies on LXR- and FXR-mediated effects on cholesterol homeostasis in normal and cholic acid-depleted mice. J. Lipid Res. 47, 421–430 [DOI] [PubMed] [Google Scholar]
- 10. Slätis K., Gåfvels M., Kannisto K., Ovchinnikova O., Paulsson-Berne G., Parini P., Jiang Z. Y., Eggertsen G. (2010) Abolished synthesis of cholic acid reduces atherosclerotic development in apolipoprotein E knockout mice. J. Lipid Res. 51, 3289–3298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ishida H., Yamashita C., Kuruta Y., Yoshida Y., Noshiro M. (2000) Insulin is a dominant suppressor of sterol 12α-hydroxylase P450 (CYP8B) expression in rat liver. Possible role of insulin in circadian rhythm of CYP8B. J. Biochem. 127, 57–64 [DOI] [PubMed] [Google Scholar]
- 12. Lavery D. J., Schibler U. (1993) Circadian transcription of the cholesterol 7α-hydroxylase gene may involve the liver-enriched bZIP protein DBP. Genes and Dev. 7, 1871–1884 [DOI] [PubMed] [Google Scholar]
- 13. Li Y. C., Wang D. P., Chiang J. Y. (1990) Regulation of cholesterol 7α-hydroxylase in the liver. Cloning, sequencing, and regulation of cholesterol 7α-hydroxylase mRNA. J. Biol. Chem. 265, 12012–12019 [PubMed] [Google Scholar]
- 14. Noshiro M., Nishimoto M., Okuda K. (1990) Rat liver cholesterol 7α-hydroxylase. Pretranslational regulation for circadian rhythm. J. Biol. Chem. 265, 10036–10041 [PubMed] [Google Scholar]
- 15. Zhang Y. K., Guo G. L., Klaassen C. D. (2011) Diurnal variations of mouse plasma and hepatic bile acid concentrations as well as expression of biosynthetic enzymes and transporters. PLoS ONE 6, e16683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Noshiro M., Usui E., Kawamoto T., Kubo H., Fujimoto K., Furukawa M., Honma S., Makishima M., Honma K., Kato Y. (2007) Multiple mechanisms regulate circadian expression of the gene for cholesterol 7α-hydroxylase (Cyp7a), a key enzyme in hepatic bile acid biosynthesis. J. Biol. Rhythms 22, 299–311 [DOI] [PubMed] [Google Scholar]
- 17. Duez H., van der Veen J. N., Duhem C., Pourcet B., Touvier T., Fontaine C., Derudas B., Baugé E., Havinga R., Bloks V. W., Wolters H., van der Sluijs F. H., Vennström B., Kuipers F., Staels B. (2008) Regulation of bile acid synthesis by the nuclear receptor Rev-erbα. Gastroenterology 135, 689–698 [DOI] [PubMed] [Google Scholar]
- 18. Crumbley C., Wang Y., Kojetin D. J., Burris T. P. (2010) Characterization of the core mammalian clock component, NPAS2, as a REV-ERBα/RORα target gene. J. Biol. Chem. 285, 35386–35392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hamilton B. A., Frankel W. N., Kerrebrock A. W., Hawkins T. L., FitzHugh W., Kusumi K., Russell L. B., Mueller K. L., van Berkel V., Birren B. W., Kruglyak L., Lander E. S. (1996) Disruption of the nuclear hormone receptor RORα in staggerer mice. Nature 379, 736–739 [DOI] [PubMed] [Google Scholar]
- 20. Kang H. S., Okamoto K., Takeda Y., Beak J. Y., Gerrish K., Bortner C. D., DeGraff L. M., Wada T., Xie W., Jetten A. M. (2011) Transcriptional profiling reveals a role for RORα in regulating gene expression in obesity-associated inflammation and hepatic steatosis. Physiol. Genomics 43, 818–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kang H. S., Angers M., Beak J. Y., Wu X., Gimble J. M., Wada T., Xie W., Collins J. B., Grissom S. F., Jetten A. M. (2007) Gene expression profiling reveals a regulatory role for RORα and RORγ in phase I and phase II metabolism. Physiol Genomics 31, 281–294 [DOI] [PubMed] [Google Scholar]
- 22. Wada T., Kang H. S., Angers M., Gong H., Bhatia S., Khadem S., Ren S., Ellis E., Strom S. C., Jetten A. M., Xie W. (2008) Identification of oxysterol 7α-hydroxylase (Cyp7b1) as a novel retinoid-related orphan receptor α (RORα) (NR1F1) target gene and a functional cross-talk between RORα and liver X receptor (NR1H3). Mol. Pharmacol. 73, 891–899 [DOI] [PubMed] [Google Scholar]
- 23. Kallen J. A., Schlaeppi J. M., Bitsch F., Geisse S., Geiser M., Delhon I., Fournier B. (2002) X-ray structure of the hRORα LBD at 1.63 Å. Structural and functional data that cholesterol or a cholesterol derivative is the natural ligand of RORα. Structure 10, 1697–1707 [DOI] [PubMed] [Google Scholar]
- 24. Kallen J., Schlaeppi J. M., Bitsch F., Delhon I., Fournier B. (2004) Crystal structure of the human RORα Ligand binding domain in complex with cholesterol sulfate at 2.2 Å. J. Biol. Chem. 279, 14033–14038 [DOI] [PubMed] [Google Scholar]
- 25. Wang Y., Kumar N., Solt L. A., Richardson T. I., Helvering L. M., Crumbley C., Garcia-Ordonez R. D., Stayrook K. R., Zhang X., Novick S., Chalmers M. J., Griffin P. R., Burris T. P. (2010) Modulation of retinoic acid receptor-related orphan receptor α and γ activity by 7-oxygenated sterol ligands. J. Biol. Chem. 285, 5013–5025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Song K. H., Chiang J. Y. (2006) Glucagon and cAMP inhibit cholesterol 7α-hydroxylase (CYP7a1) gene expression in human hepatocytes. Discordant regulation of bile acid synthesis and gluconeogenesis. Hepatology 43, 117–125 [DOI] [PubMed] [Google Scholar]
- 27. Li T., Francl J. M., Boehme S., Ochoa A., Zhang Y., Klaassen C. D., Erickson S. K., Chiang J. Y. (2012) Glucose and Insulin Induction of Bile Acid Synthesis. Mechanisms and implication in diabetes and obesity. J. Biol. Chem. 287, 1861–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang M., Chiang J. Y. (2001) Transcriptional regulation of the human sterol 12α-hydroxylase gene (CYP8B1). Roles of hepatocyte nuclear factor 4α (HNF4α) in mediating bile acid repression. J. Biol. Chem. 276, 41690–41699 [DOI] [PubMed] [Google Scholar]
- 29. Han S., Chiang J. Y. (2009) Mechanism of vitamin D receptor inhibition of cholesterol 7α-hydroxylase gene transcription in human hepatocytes. Drug Metab. Dispos. 37, 469–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li T., Chiang J. Y. (2007) A novel role of transforming growth factor β1 in transcriptional repression of human cholesterol 7α-hydroxylase gene. Gastroenterology 133, 1660–1669 [DOI] [PubMed] [Google Scholar]
- 31. Han S., Li T., Ellis E., Strom S., Chiang J. Y. (2010) A novel bile acid-activated vitamin D receptor signaling in human hepatocytes. Mol. Endocrinol. 24, 1151–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Belle A., Tanay A., Bitincka L., Shamir R., O'Shea E. K. (2006) Quantification of protein half-lives in the budding yeast proteome. Proc. Natl. Acad. Sci. U.S.A. 103, 13004–13009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang Y., Klaassen C. D. (2010) Effects of feeding bile acids and a bile acid sequestrant on hepatic bile acid composition in mice. J. Lipid Res. 51, 3230–3242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jetten A. M. (2009) Retinoid-related orphan receptors (RORs). Critical roles in development, immunity, circadian rhythm, and cellular metabolism. Nucl. Recept. Signal. 7, e003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Feng D., Liu T., Sun Z., Bugge A., Mullican S. E., Alenghat T., Liu X. S., Lazar M. A. (2011) A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science 331, 1315–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Brufau G., Stellaard F., Prado K., Bloks V. W., Jonkers E., Boverhof R., Kuipers F., Murphy E. J. (2010) Improved glycemic control with colesevelam treatment in patients with type 2 diabetes is not directly associated with changes in bile acid metabolism. Hepatology 52, 1455–1464 [DOI] [PubMed] [Google Scholar]
- 37. Haeusler R. A., Pratt-Hyatt M., Welch C. L., Klaassen C. D., Accili D. (2012) Impaired generation of 12-hydroxylated bile acids links hepatic insulin signaling with dyslipidemia. Cell Metab. 15, 65–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Puigserver P., Rhee J., Donovan J., Walkey C. J., Yoon J. C., Oriente F., Kitamura Y., Altomonte J., Dong H., Accili D., Spiegelman B. M. (2003) Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1α interaction. Nature 423, 550–555 [DOI] [PubMed] [Google Scholar]
- 39. Nakae J., Kitamura T., Silver D. L., Accili D. (2001) The forkhead transcription factor Foxo1 (Fkhr) confers insulin sensitivity onto glucose-6-phosphatase expression. J. Clin. Invest. 108, 1359–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li T., Kong X., Owsley E., Ellis E., Strom S., Chiang J. Y. (2006) Insulin regulation of cholesterol 7α-hydroxylase expression in human hepatocytes. Roles of forkhead box O1 and sterol regulatory element-binding protein 1c. J. Biol. Chem. 281, 28745–28754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li T., Ma H., Park Y. J., Lee Y. K., Strom S., Moore D. D., Chiang J. Y. (2009) Forkhead box transcription factor O1 inhibits cholesterol 7α-hydroxylase in human hepatocytes and in high fat diet-fed mice. Biochim. Biophys. Acta 1791, 991–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li T., Owsley E., Matozel M., Hsu P., Novak C. M., Chiang J. Y. (2010) Transgenic expression of cholesterol 7α-hydroxylase in the liver prevents high-fat diet-induced obesity and insulin resistance in mice. Hepatology 52, 678–690 [DOI] [PMC free article] [PubMed] [Google Scholar]