Abstract
While reversible protein phosphorylation plays an important role in many cellular processes, simple and reliable measurement of the stoichiometry of phosphorylation can be challenging. This measurement is confounded by differences in the ionization efficiency of phosphorylated and unphosphorylated sites during analysis by mass spectrometry. Here, we demonstrate diagonal capillary electrophoresis-mass spectrometry for the accurate determination of this stoichiometry. Diagonal capillary electrophoresis is a two-dimensional separation method that incorporates an immobilized alkaline phosphatase microreactor at the distal end of the first capillary and employs identical electrophoretic separation modes in both dimensions. The first dimension is used to separate a mixture of the phosphorylated and unphosphorylated forms of a peptide. Fractions are parked in the reactor where they undergo complete dephosphorylation. The products are then periodically transferred to the second capillary and analyzed by mass spectrometry (MS). Because the phosphorylated and unphosphorylated forms differ in charge, they are well resolved in the first dimension separation. Because the unphosphorylated and dephosphorylated peptides are identical, there is no bias in ionization efficiency, and phosphorylation stoichiometry can be determined by the ratio of the signal of the two forms. A calibration curve was generated from mixtures of a phosphorylated standard peptide and its unphosphorylated form, prepared in a bovine serum albumin tryptic digest. This proof of principle experiment demonstrated a linear response across nearly two orders of magnitude in stoichiometry.
Reversible protein phosphorylation plays an important role in regulating a wide range of cellular processes, including proliferation, differentiation, transformation, cell cycle control, receptor-mediated signal transduction, and metabolism.1–4 Mass spectrometry-based proteomic analyses have generated large-scale maps of phosphorylation sites.5,6 Protocols incorporating stable-isotope labeling with mass spectrometry have produced datasets that illustrate protein phosphorylation changes under a variety of conditions.7–12 There often is ambiguity in these data; the change in the abundance in a phosphorylated protein can be due either to a change in the overall expression of that protein with constant phosphorylation stoichiometry or to a change in the phosphorylation stoichiometry with constant overall protein expression.13
Phosphorylation stoichiometries can be quantified by dividing the signal for a given phosphopeptide by the sum of signals of its phosphorylated and unphosphorylated forms. This procedure often results in an underestimation of the phosphorylation stoichiometry because phosphopeptides ionize with lower efficiency than their corresponding unphosphorylated form.14,15 Alternatively, the sample can be analyzed before and after treatment with alkaline phosphatase, which quantitatively removes phosphate groups.16–17 This approach requires use of isotopic labels and corresponding sample manipulations, and is best suited for characterization of the stoichiometry of a very large number of phosphorylated peptides in a small number of samples.
In this manuscript, we present a proof-of-principle demonstration of an unbiased method for determining phosphorylation stoichiometries that may be of value for high-throughput characterization of a relatively small number of phosphorylated peptides. Our method is based on a diagonal separation. Diagonal separations have been used for characterization of post-translational modifications since the earliest example of screening disulfide bonds in peptides using paper chromatography.18,19 In diagonal separations, identical separation modes are employed in both dimensions of a two-dimensional separation. A treatment is employed between dimensions. Analytes that are unchanged by the treatment will have identical properties in both dimensions, which produces a separation consisting of a set of spots that form a diagonal. Those components that undergo modification between separations will generate off-diagonal spots.20 Diagonal technology has evolved into diagonal liquid chromatography and diagonal gel electrophoresis for study of post-translational modifications.21–23
We reported a diagonal capillary electrophoresis method for characterization of peptide phosphorylation.20,24 That method employed a microreactor consisting of alkaline phosphatase that was immobilized at the distal end of the first capillary of the two-dimensional system. A mixture of peptides was introduced into the first capillary and separated based on zone electrophoresis. Fractions from this capillary were parked in the microreactor. After reaction, those fractions were transferred to the second capillary, where the components were again separated by zone electrophoresis under the same conditions. Unphosphorylated peptides that passed through the reactor without change had identical mobility in the first and second separation dimensions, and those components formed a diagonal in the reconstructed electropherogram. In contrast, phosphorylated peptides were dephosphorylated in the reactor and appeared off the diagonal.
The extent of dephosphorylation produced by the microreactor can be tuned by adjusting the length of the reactor, the amount of immobilized enzyme, the incubation time, and temperature. In our original descriptions of the technique,20,24 we employed conditions that lead to incomplete dephosphorylation, so that both the phosphorylated and dephosphorylated forms were present, which allowed us to identify phosphorylation sites. Our most recent publication used reactor conditions that incompletely removed phosphate groups. We employed this reactor in diagonal capillary electrophoresis coupled with an LTQ-Orbitrap Velos mass spectrometer to characterize the phosphorylation status of a tryptic digest of α-casein in a 22-fold excess of the tryptic digest of bovine serum albumin. Nine phosphorylated α-casein peptides that produced 20 different phosphorylation states were detected with high confidence24.
In this study, we employed conditions that produced complete dephosphorylation. These conditions are ideal for the determination of phosphorylation stoichiometry. Diagonal electrophoresis first separates the unphosphorylated and phosphorylated forms of the peptide, and then converts the phosphorylated peptide into its dephosphorylated form. The unphosphorylated and dephosphorylated peptides are identical, as are their ionization efficiencies. When dephosphorylation is complete, the ratio of the intensities of the unphosphorylated and dephosphorylated peptides provides an accurate measure of phosphorylation status of the peptide. This manuscript provides a proof-of-principle demonstration of the technique.
EXPERIMENTAL SECTION
Materials and Chemicals
Acrylamide (AA), ammonium persulfate (APS), tetramethylethylenediamine (TEMED), ammonium bicarbonate (NH4HCO3), N, N’-methylenebisacrylamide (MBA), acrylic acid N-hydroxyl-succinimide ester (NAS), 3-(trimethroxysilyl)propyl methacrylate, dithiothreitol (DTT), iodoacetamide (IAA), alkaline phosphatase from bovine intestinal mucosa, bovine serum albumin (BSA), TPCK-treated trypsin were from Sigma Aldrich (St. Louis, MO, USA). Formic acid (FA) was from Fisher Scientific (Pittsburgh, PA, USA). Water and methanol were from Honeywell Burdick & Jackson (Muskegon, MI, USA). Fused silica capillaries were from Polymicro Technologies (Phoenix, AZ, USA). ZipTipC18 was from Millipore Co. (Gilbert, AZ, USA).
Sample
The commercially available mono-phosphorylated (m/z 852, z = +2, H - Thr - Arg - Asp - Ile - pTyr - Glu - Thr - Asp - Tyr - Tyr - Arg - Lys - OH) and unphosphorylated (m/z 812, z = +2, H - Thr - Arg - Asp - Ile - Tyr - Glu - Thr - Asp - Tyr - Tyr - Arg - Lys - OH) standard peptides were derived from insulin receptor (AnaSpec, Fremont, CA, USA).
The BSA digest was prepared as previously reported.25 Briefly, 8 µL of 1 M DTT was added to 1 mL of 1 mg/mL BSA in 8 M urea and 100 mM NH4HCO3 (pH 8.0). The solution was kept at 60 °C for 1 h for reduction. 20 µL of 1 M IAA was added to the solution and incubated at room temperature for 30 min in the dark for alkylation. After four-times dilution to reduce the urea concentration to 2 M, TPCK-treated trypsin digestion was performed at 37 °C overnight with a trypsin/protein ratio of 1:30 (w/w). The digests were acidified and desalted with C18 SepPak cartridges from Waters (Milford, MA, USA) and lyophilized, followed by analysis using diagonal capillary electrophoresis.
The sample was prepared by mixing the unphosphorylated peptide (m/z 812, z = +2, 100 µM in separation buffer) and the mono-phosphorylated (m/z 852, z = +2, 100 µM in separation buffer) in molar ratios of 5:95, 3:7, 30:70, and 70:30, with 0.5 mg/mL BSA digest as background.
Preparation of the open-tubular alkaline phosphatase-immobilized microreactor
A fused silica capillary (48 µm i.d./148 µm o.d.) was flushed with methanol (20 min), N2 gas (10 min), 1 M NaOH (20 min), H2O (10 min), 1 M HCl (20 min), H2O (10 min), methanol (20 min), and N2 gas (20 min). A portion of the treated capillary (~10 cm) was filled with a mixture of methanol and 3-(trimethroxysilyl) propyl methacrylate (1:1 (v/v)) by capillary action, and the reaction was allowed to proceed at room temperature for 24 h. The capillary was then washed with methanol to remove the unreacted components, and dried again with nitrogen. The treated capillary was stored at room temperature before use.
The general protocol of reference 25 was used for monolith polymerization and enzyme immobilization, with the following modifications. A mixture of 2.5 mg of AA, 3.75 mg of MBA, and 30 mg of PEG was dissolved in 1 mL of 0.2 M sodium bicarbonate/0.5 M sodium chloride (pH ~8.0) buffer. The mixture was vortexed for 30 s and then heated at ~50 °C for 15 min to completely dissolve the monomers. Then, 2 µL of 20% (v/v) TEMED was added into 0.5 mL of the prepared mixture, and the mixture was degassed for 15 min using nitrogen. Subsequently, 7 µL of NAS (140 mg/mL, dissolved in DMSO) was added on top of the solution, and degassed for 1 min. Next, 2 µL of 20% (w/v) APS was added into the middle part of the solution to initiate polymerization. After vortexing for several seconds, a 95 µL-aliquot of the solution was quickly mixed with 5 µL of alkaline phosphatase from bovine intestinal mucosa (13 mg/mL in a buffer containing Mg2+ and Zn2+). The activated portion of the treated capillary (~8 cm in length) was filled with the mixed solution, and reacted for 1 h at room temperature with the capillary ends sealed. After reaction, the capillary was successively washed with deionized water, glycine (10 mg/mL in phosphate buffer, pH 8.0), and 5 mM NH4HCO3 (pH ~8.0) for 30 min. The capillary was stored at 4 °C before use. The capillary was trimmed to the desired length before use. Typically, the overall capillary length was 28 cm, and the reactor occupied 4~5 cm at the distal end of the capillary.
Diagonal CE-ESI-MS/MS Analysis
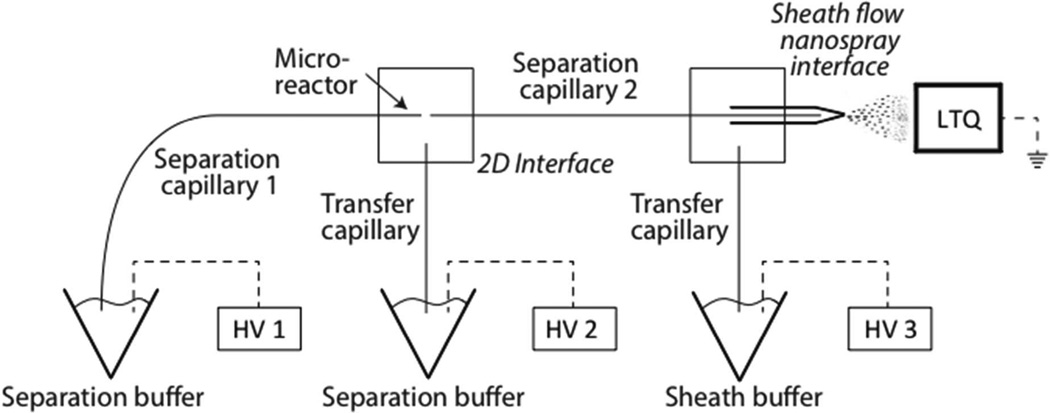
The diagonal capillary electrophoresis system was coupled to a mass spectrometer using an electrokinetically driven sheath-flow nanospray interface.27–29 The system has been described previously and is shown in Figure 1.19,23 Two capillaries (20 µm i.d./136 µm o.d., 25 cm length) were connected through a nicked-gap interface.30 The capillary spacing was adjusted to ~20-µm. The interface was filled with the separation buffer. High voltage power supplies were connected to the injection and interface buffer vials, and to the nanospray emitter.
Figure 1.
Schematic diagram of the diagonal capillary electrophoresis system. An LTQ-XL mass spectrometer was connected to the electrophoresis system through the electrokinetically-pumped sheath-flow nanospray interface. A ~4.5 cm open-tube monolithic microreactor was synthesized at the distal end of the first capillary.
Sample injection was performed by applying 10 kV to power supply 1, 5 kV to power supply 2, and 0 kV to power supply 3 for one second. The sample was replaced with separation buffer, and a preliminary separation was performed (19.5 kV to power supply 1, 10.5 kV to power supply 2, and 1.5 kV to power supply 3).31 The mass spectrometer was held at ground potential. The time for the initial separation was half of the sample’s migration time in a pseudo-one dimensional capillary electrophoresis separation, where the same electric field was continuously applied across both capillaries.
Cycles of fraction transfers and second dimension separations were then started. Each cycle lasted for 52 seconds. The two-second duration transfers were driven by the same potentials that were used in the preliminary separation. The 50-s duration second-dimension separations were performed by applying a potential of 10.5 kV to power supplies 1 and 2, which resulted in a zero volt potential across capillary 1, and 1.5 kV to power supply 3. The 50-s duration of the second dimension separation was sufficiently long to produce complete dephosphorylation of components present in the reactor.
An LTQ-XL instrument (Thermo Fisher Scientific) was used for peptide detection. Full MS scans were acquired in the linear ion trap mass analyzer over the m/z 350–1800 range.
RESULTS AND DISCUSSION
Diagonal capillary electrophoresis analysis
We begin by defining nomenclature. The unphosphorylated form of the peptide contains no phosphate groups. The parent phosphorylated form of the peptide contains one or more phosphate groups and undergoes dephosphorylation in the microreactor to produce the dephosphorylated form. The unphosphorylated and dephosphorylated forms are identical molecules with identical mobility in the second separation dimension and in ionization efficiency.
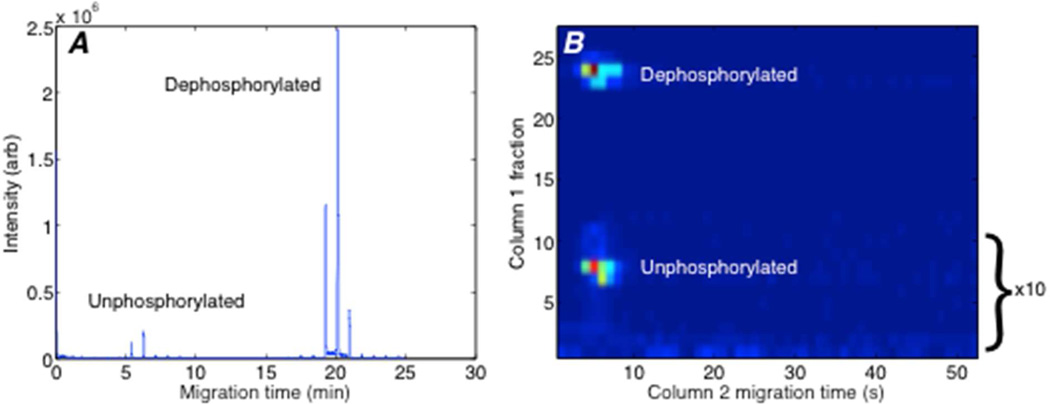
Figure 2 presents the selected-ion electropherogram generated from a mixture of standard peptides, 95% of which were phosphorylated, in 0.5 mg/mL tryptic bovine serum albumin digest. The high activity reactor completely dephosphorylated peptides. As a result, only the unphosphrylated and dephosphorylated forms were observed for the standard peptides, and the parent, mono-phosphorylated form of the peptide (TRDIVETD-pY-YRK, m/z 852, z = +2) was not detected. Instead, the phosphorylated peptide was quantitatively converted to the dephosphorylated form (m/z 812, z = +2). The time trace consists of a set of peaks for each peptide, and those peaks are separated by 52 seconds, which is the second-dimension cycle time. Successive transfers to the second capillary generated two or three peaks from each peptide.
Figure 2.
Two-dimensional capillary electrophoresis analysis of an unphosphorylated standard peptide (TRDIVETDYYRK) and the mono-phosphopeptide (TRDIVETD-pY-YRK), which is converted to the dephosphorylated form. A. Raw, unprocessed data. B. Reconstructed two-dimensional electropherogram. The intensity of the first 12 transfers was multiplied by 10 to bring the signal from the unphosphorylated peptide onto a similar scale as the dephosphorylated peptide. A two-dimensional Gaussian surface was fit to each of the spots. The spot widths for the unphosphorylated spot were 0.60 transfers in the y dimension and 1.3 s in the x dimension. The spot widths for the dephosphorylated spot were 0.51 transfers in the y dimension and 1.1 s in the x dimension.
The reconstructed two-dimensional separation is shown in Figure 2B. The separation was performed in an uncoated capillary at basic pH. At this pH, the electro-osmotic mobility of unphosphorylated peptides was higher than the electrophoretic mobility of the phosphorylated peptides, which carry additional negative charge and migrate more slowly than unphosphorylated peptides. The migration time of the dephosphorylated peptide in the first dimension separation is determined by the migration time of the parent, phosphorylated peptide from which it is formed. Based on the width of the envelope, the parent mono-phosphate form of the peptide had a ~1.2 s width in the first capillary. The unphosphorylated and dephosphorylated peptides have identical migration time in the second dimension because they are identical molecules.
The peptide signal was calculated by adding peak areas of each set of peptide (Peaks with signal-to-noise ratio less than 100 were excluded). Peak areas were obtained by Xcalibur software from the raw file. Phosphorylation stoichiometry was estimated by dividing integrated peak signal for the dephosphorylated form of a given phosphopeptide by the sum of signals of its dephosphorylated and unphosphorylated forms. All phosphopeptides were completely dephosphorylated in the reactor. In Figure 2, the area ratio between dephosphopeptide and the total standard signal was 0.92, compared with the theoretical stoichiometry of 0.95.
Calibration curve
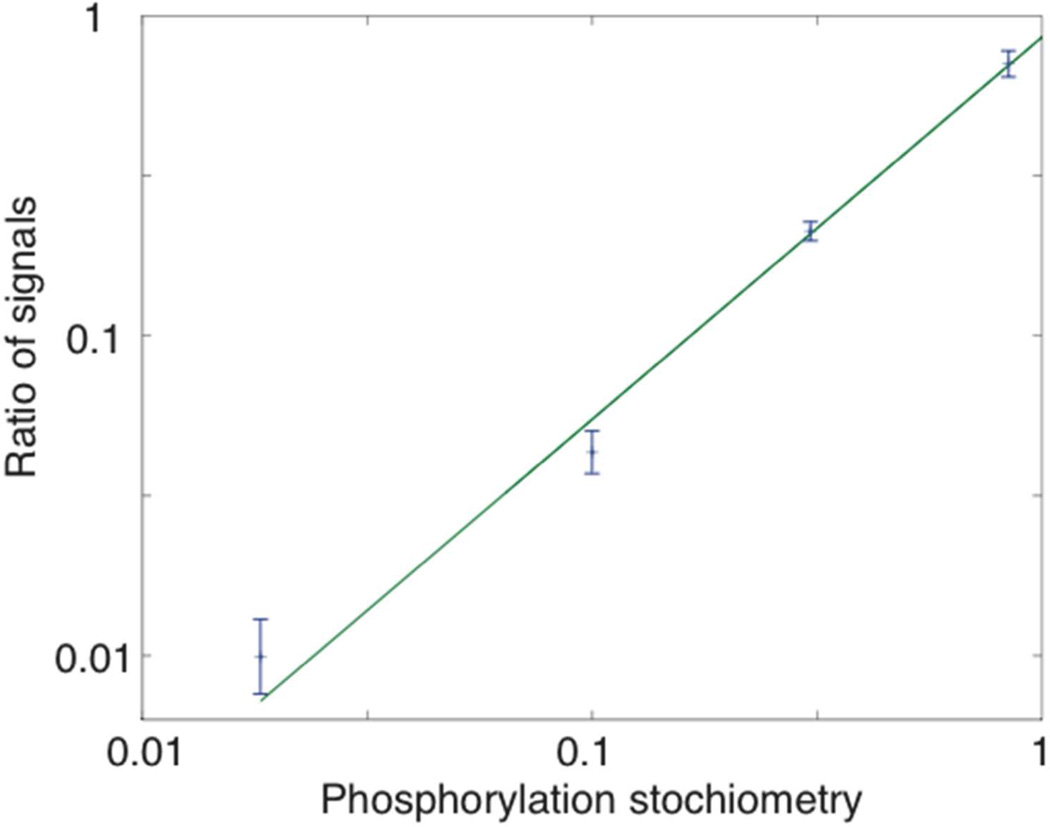
The unphosphorylated peptide (TRDIVETDYYRK) and its mono-phosphorylated form were mixed in ratios of 5:95, 30:70, 50:50 and 70:30, producing phosphorylation stoichiometries of 95%, 70%, 50% and 30%, respectively. Each was mixed with 0.5 mg/mL BSA tryptic digests to mimic a real-world analysis. A calibration curve was generated from the peak area of dephosphorylated form over total peptide standard. Three replicates were obtained at each ratio. The stoichiometries spanned nearly two orders of magnitude, which should be sufficient to monitor the change in phosphorylation status of proteins in the signaling networks.32
A log-log plot of the calibration curve generated a slope of 1.19, which confirms the data’s linearity. Table 1 shows the results of the regression analysis. The mean RSD of peak area is 4% demonstrating the accuracy and reproducibility in peak area in a contextual background.
Table 1.
Results of a linear fit to the log-log calibration curve (Figure 3)
| Slope | χ2 | R2 | mean RSD peak area |
|---|---|---|---|
| 1.19 ± 0.06 | 3.8 (p>0.15) | 0.994 | 4% |
The alkaline phosphatase microreactor can be stored in the separation buffer at 4 °C for at least one month without significant loss of enzymatic activity. We usually run more than ten experiments per day using the microreactor. The reactor maintains high activity over at least one week under this constant use. Furthermore, when coupled with multiple-reaction monitoring,33–34 it should be possible to characterize the stoichiometry of dozens of sites. This performance suggests that the technique can be used for the high-throughput determination of phosphorylation stoichiometry for a modest number of sites.
Figure 3.
Log-log calibration curve for unphosphorylated and mono-phosphorylated standards with different phosphorylation stoichiometries. Data are plotted as ± 1 standard deviation of the mean, n = 3 for each stoichiometry. The smooth curve is the weighted least squares fit of a linear polynomial to the logarithm of the data. Results of the fit are summarized in Table 1.
ACKNOWLEDGEMENTS
We thank Dr. William Boggess of the Notre Dame Mass Spectrometry and Proteomics Facility for his help with this project. This work was supported by a grant from the National Institutes of Health (R01GM096767).
Footnotes
The authors declare no competing financial interest.
REFERENCES
- 1.Macek B, Mann M, Olsen JV. Annu. Rev. Pharmacol. Toxicol. 2009;49:199–211. doi: 10.1146/annurev.pharmtox.011008.145606. [DOI] [PubMed] [Google Scholar]
- 2.Cohen P. Nat. Cell Biol. 2002;4:127–130. doi: 10.1038/ncb0502-e127. [DOI] [PubMed] [Google Scholar]
- 3.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 4.Wu R, Dephoure N, Haas W, Huttlin EL, Zhai B, Sowa ME, Gygi SP. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.M111.009654. M111.009654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bensimon A, Heck AJ, Aebersold R. Annu. Rev. Biochem. 2012;81:379–405. doi: 10.1146/annurev-biochem-072909-100424. [DOI] [PubMed] [Google Scholar]
- 6.Mann M, Ong SE, Gronborg M, Steen H, Jensen ON, Pandey A. Trends Biotechnol. 2002;20:261–268. doi: 10.1016/s0167-7799(02)01944-3. [DOI] [PubMed] [Google Scholar]
- 7.Ong SE, Mann M. Nat. Protoc. 2007;1:2650–2660. doi: 10.1038/nprot.2006.427. [DOI] [PubMed] [Google Scholar]
- 8.Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ. Mol. Cell. Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Thompson A, Schafer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, Neumann T, Johnstone R, Mohammed AK, Hamon C. Anal. Chem. 2003;75:1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- 10.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Nat. Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 11.Alghamdi WM, Gaskell SJ, Barber J. Anal. Chem. 2012;84:7384–7392. doi: 10.1021/ac301038u. [DOI] [PubMed] [Google Scholar]
- 12.Broberg A. Carbohydr. Res. 2007;342:1462–1469. doi: 10.1016/j.carres.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 13.Kanshin E, Michnick S, Thibault P. Semin Cell Dev Biol. 2012;23:843–853. doi: 10.1016/j.semcdb.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 14.Radimerski T, Mini T, Schneider U, Wettenhall RE, Thomas G, Jenoe P. Biochemistry. 2000;39:5766–5774. doi: 10.1021/bi9927484. [DOI] [PubMed] [Google Scholar]
- 15.Resing KA, Ahn NG. Methods Enzymol. 1997;283:29–44. doi: 10.1016/s0076-6879(97)83005-4. [DOI] [PubMed] [Google Scholar]
- 16.Domanski D, Murphy LC, Borchers CH. Anal. Chem. 2010;82:5610–5620. doi: 10.1021/ac1005553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu R, Haas W, Dephoure N, Huttlin EL, Zhai B, Sowa ME, Gygi SP. Nat. Methods. 2011;8:677–683. doi: 10.1038/nmeth.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown JR, Hartley BS. Biochem. J. 1963;89:59–60. [Google Scholar]
- 19.Brown JR, Hartley BS. Biochem. J. 1966;101:214–228. doi: 10.1042/bj1010214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wojcik R, Vannatta M, Dovichi NJ. Anal. Chem. 2010;82:1564–1567. doi: 10.1021/ac100029u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gevaert K, Van Damme J, Goethals M, Thomas GR, Hoorelbeke B, Demol H, Martens L, Puype M, Staes A, Vandekerckhove J. Mol. Cell. Proteomics. 2002;1:896–903. doi: 10.1074/mcp.m200061-mcp200. [DOI] [PubMed] [Google Scholar]
- 22.Chernov IP, Timchenko KA, Akopov SB, Nikolaev LG, Sverdlov ED. Anal. Biochem. 2007;364:60–66. doi: 10.1016/j.ab.2007.01.040. [DOI] [PubMed] [Google Scholar]
- 23.Chernov IP, Akopov SB, Nikolaev LG, Sverdlov ED. Biotechniques. 2006;41:91–96. [PubMed] [Google Scholar]
- 24.Mou S, Sun L, Wojcik R, Dovichi NJ. Talanta. 2013;116:985–990. doi: 10.1016/j.talanta.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun L, Li Y, Yang P, Zhu G, Dovichi NJ. J. Chromatogr. A. 2012;1220:68–74. doi: 10.1016/j.chroma.2011.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palm AK, Novotny MV. Rapid Commun. Mass Spectrom. 2004;18:1374–1382. doi: 10.1002/rcm.1500. [DOI] [PubMed] [Google Scholar]
- 27.Wojcik R, Dada OO, Sadilek M, Dovichi NJ. Rapid Commun. Mass Spectrom. 2010;24:2554–2560. doi: 10.1002/rcm.4672. [DOI] [PubMed] [Google Scholar]
- 28.Sun L, Zhu G, Li Y, Wojcik R, Yang P, Dovichi NJ. Proteomics. 2012;12:3013–3019. doi: 10.1002/pmic.201200100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y, Champion MM, Sun L, Champion PAD, Wojcik R, Dovichi NJ. Anal. Chem. 2012;84:1617–1622. doi: 10.1021/ac202899p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flaherty RJ, Huge BJ, Bruce SM, Dada OO, Dovichi NJ. Analyst. 2013;138:3621–3625. doi: 10.1039/c3an00284e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schoenherr RM, Ye M, Vannatta M, Dovichi NJ. Anal. Chem. 2007;79:2230–2238. doi: 10.1021/ac061638h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 33.Sun L, Li Y, Champion MM, Zhu G, Wojcik R, Dovichi NJ. Analyst. 2013;138:3181–3188. doi: 10.1039/c3an00287j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y, Wojcik R, Dovichi NJ, Champion MM. Anal. Chem. 2012;84:6116–6121. doi: 10.1021/ac300926h. [DOI] [PMC free article] [PubMed] [Google Scholar]