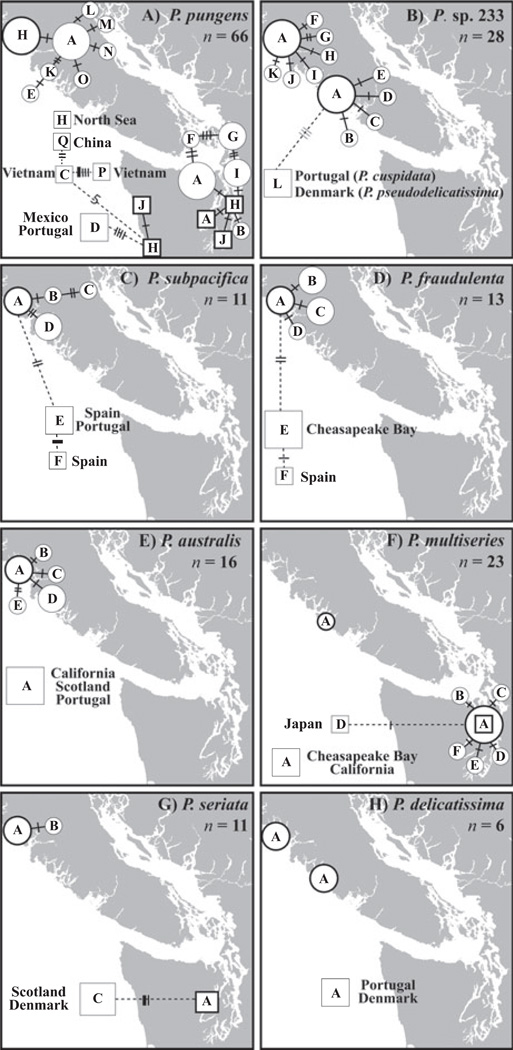
Fig. 3.
Network of polymorphic genotypes for eight Pseudo-nitzschia species/species types: (A) P. pungens; (B) P. sp. 233; (C) P. subpacifica; (D) P. fraudulenta; (E) P. australis; (F) P. multiseries; (G) P. seriata; and (H) P. delicatissima type 11. ITS sequences from clone libraries (represented by circles) generated in this study and from isolates from GenBank and this study (represented by squares) were used. The size of the circle or square represents the number of sequences; the smallest size represents a single sequence, the medium size represents two to four sequences, and the largest size represents five or more sequences. Unique genotypes are distinguished by letters located inside circles or squares; thickly outlined circles and squares, or “hubs,” are placed in the actual location where the sample or isolate originated. Solid branches are used to connect genotypes from the Pacific Northwest; dotted branches are used to connect genotypes from other regions. Crossbars represent differences between genotypes; thin bars represent deletions relative to the hub sequence, medium bars represent a polymorphism, and thick bars represent insertions relative to the hub sequence. ITS, internal transcribed spacer.