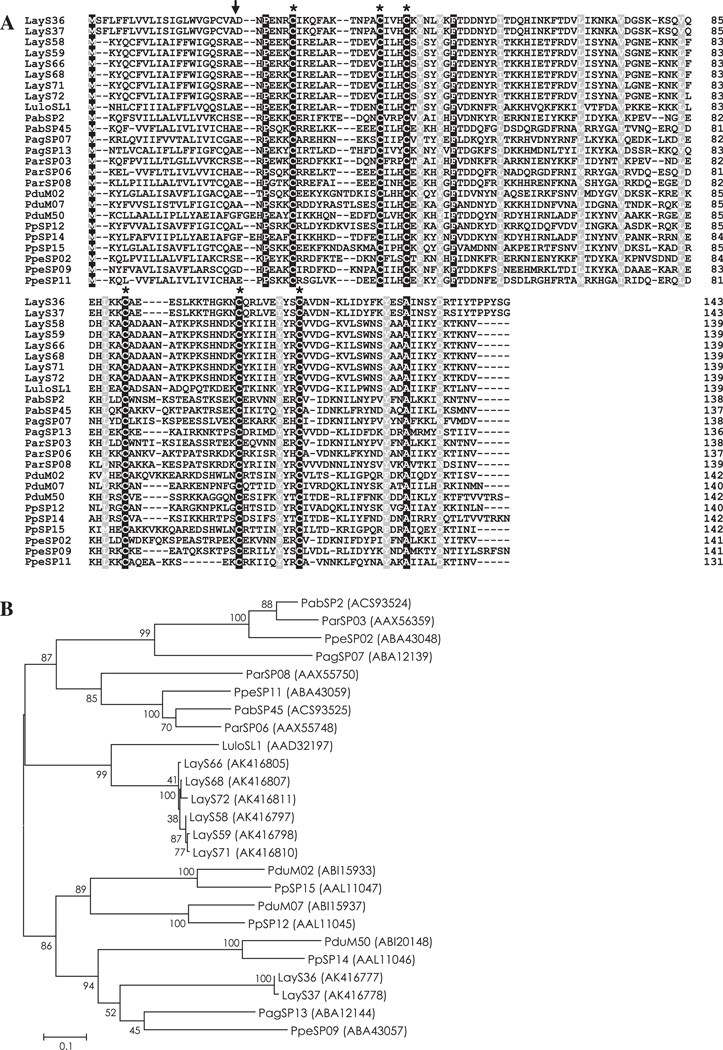
Fig. 1.
(A) Sequence alignment of PpSP15/SL1 family proteins from Lutzomyia (Lu.) ayacuchensis (LayS36, LayS37, LayS58, LayS59, LayS66, LayS68, LayS71, and LayS72) together with those from Phlebotomus (P.) arabicus (Pab), P. argentipes (Pag), P. ariasi (Par), P. duboscqi (Pdu), P. papatasi (Pp), P. perniciosus (Ppe), and Lu. longipalpis (Lulo). Black-shaded amino acids represent identical amino acids and gray-shaded amino acids represent conserved amino acids. Dashes indicate gaps introduced for maximal alignment. Asterisks at the top of the amino acids denote conserved cysteine residues, and an arrowhead indicates the predicted signal peptide cleavage site. (B) Phylogenetic tree analysis of amino acid sequences of PpSP15/SL1 family proteins from sand flies. Accession numbers are in parentheses and node values indicate branch support.