Figure 2. SwrA is bound to PA(fla/che) through DegU~P.
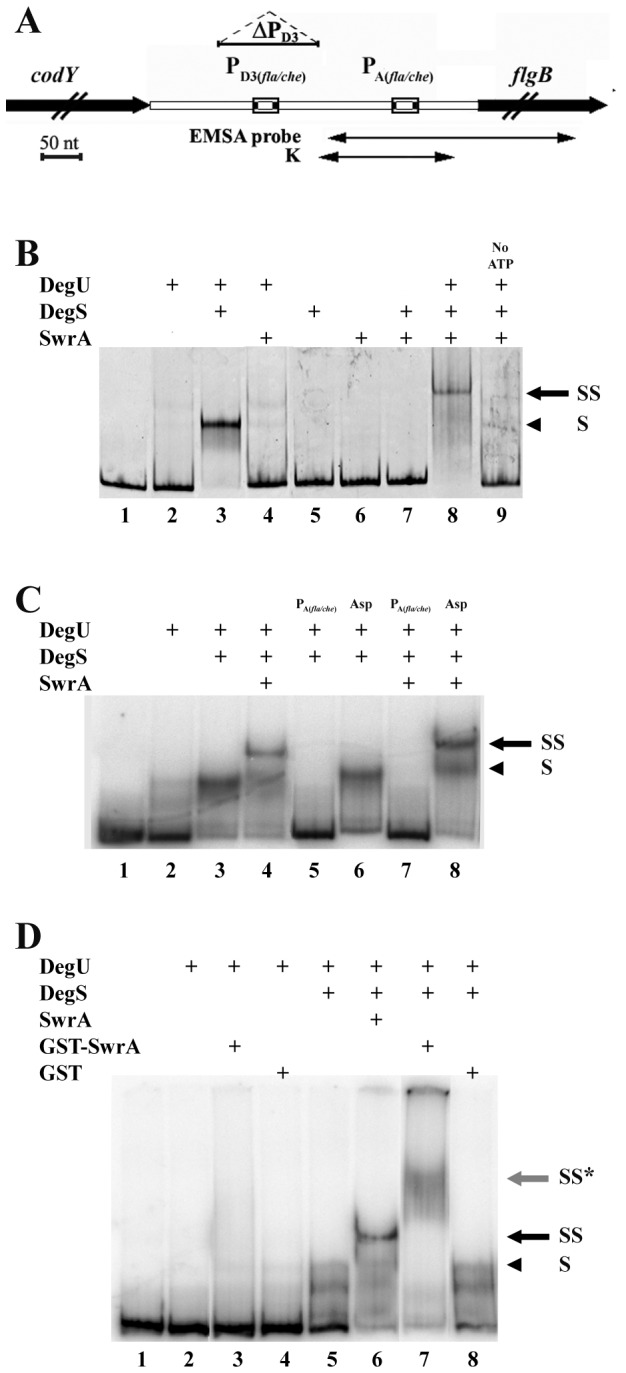
(A) The region of the fla/che promoter, including the σA and σD-dependent promoters, is drawn to scale. The coding sequences of the preceding gene (codY) and of the first gene of fla/che (flgB) are in black. The span of the PA(fla/che) probe, of the K probe used in Figure S1 and of the markerless PD3 deletion are indicated. The bar corresponds to 50 nt. (B) EMSA with fluorescently labeled PA(fla/che) probe. DegU, DegS and SwrA (listed on the left above the gel) were added to reactions labeled with a + above the lane. In each 10 μL-reaction proteins had the following concentrations: 0.35 μM for DegU; 0.4 μM for DegS; 1 μM for SwrA. ATP was omitted from the reaction in lane 9. On the right of the gel, the arrowhead labeled with S points to the probe shifted by DegU~P (lane 3); the SS arrow marks the super-shifted complex formed upon SwrA addition (lane 8). (C) Competition reactions with an excess (20 ng) of unlabeled PA(fla/che) DNA or with the same amount of unlabeled non-specific (N-S) DNA are indicated above the gel. The EMSA probe was radioactively labeled. In each reaction, the proteins had following concentration: 0.2 μM DegU, 0.1 μM DegS and 0.2 μM SwrA. (D) A GST-tag fused to SwrA reduces the migration of the SS complex. Radioactively labeled PA(fla/che) was incubated with DegU (0.1 μM), DegS (0.1 μM), and either SwrA (lane 6), GST-SwrA (lane 7) or GST (lane 8), each at 1.5 μM. The grey arrow labeled with SS* points at the retarded band obtained with GTS-SwrA (lane 7).
