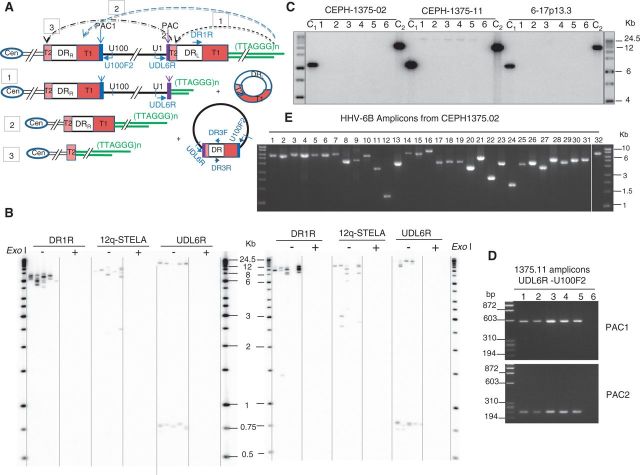
Figure 4.
The integrated viral genome can be truncated, leading to the formation of short telomeres at new locations and to the release of circular DNA molecules that contain viral sequences. (A) Truncation events [1–3] that could arise via t-loop formation between the telomeric 3' single-strand overhang and viral T1 or T2 regions and the circular molecules that could be released. Truncation at DRL-T2 [1] could arise through t-loop formation between the telomere and DRL-T2, and small circular molecules comprising one DR region could be released. T-loop formation between the telomere and the internal DRR-T1 [2] or between a chromosome that had already undergone a truncation at DRL-T2 and the internal DRR-T2 [3] could be cleaved to release a large circular molecule composed of the entire viral genome and a single DR region reconstituted with both PAC1 and PAC2. The reciprocal product would be a chromosome that retains a single DR (without PAC1 or PAC2) plus a telomere [2] or the complete removal of the viral sequence from the chromosome [3]. (B) Detection of CI-HHV-6 molecules truncated at DRL-T2 in two CI-HHV-6B carriers [CEPH1375.02, left and 4-11p15.5 (24), right]. Telomeres were detected at the end of the full-length virus using primer DR1R and at 12q (12q-STELA), as expected. The amplicons generated from a primer located near the U1 gene (UDL6R) fell into two clusters. Amplicons ∼750 bp in length were derived from truncated CI-HHV-6 molecules with short telomeres at DRL-T2. The larger products generated with the UDL6R primer are equivalent to the products from the DR1R primer. STELA products were only seen in the DNA without ExoI digestion, showing they are dependent on the presence of a telomeric 3' single-strand overhang. (C) Detection of molecules that encompass the HHV-6B unique region and a single DR with both PAC1 and PAC2. Amplification of multiple aliquots of genomic DNA (90 ng/reaction) from CEPH1375.11 (grandfather) using the primers UDL6R and U100F2 (shown in A) that are only directed towards one another on circular DNA molecules, generated large amplicons in five of the six PCRs shown but not from other CI-HHV-6B carriers in the same family (CEPH 1375.02, mother shown) or in an unrelated CI-HHV-6B carrier (6-17p13.3). Control amplicons, C1 (UDL6R + DR3F) and C2 (U100F2 + DR3R), were generated in all the samples. The amplicons were detected by hybridization to a probe for DR3. (D) The amplicons generated from circular HHV-6 DNA molecules in CEPH1375.11 contain a single DR with both PAC1 and PAC2. Specific amplifications show the presence of PAC1 and PAC2 in each amplicon generated in (C). (E) The complete HHV-6B genome is present in the CEPH1375 family. Overlapping amplicons (1–31) covering the unique portion of the HHV-6B genome and the DRs (amplicon 32, without T1 and T2) were generated from CEPH1375.02. Each amplicon is the expected size showing there are no gross rearrangements of the integrated viral genome.