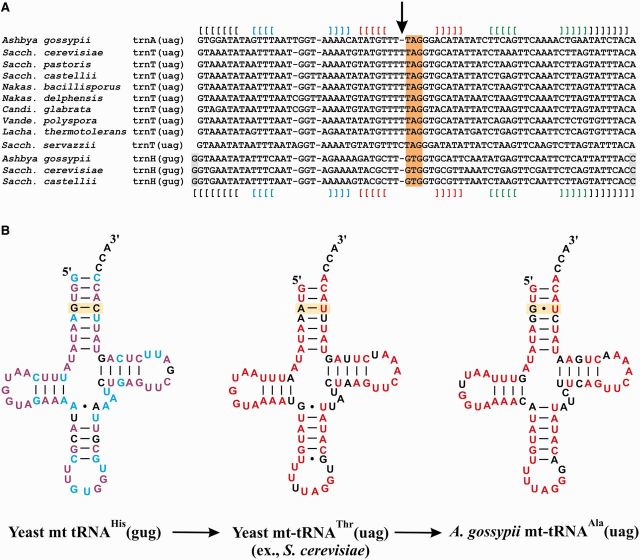
Figure 2.
Primary and secondary structures of yeast mitochondrial 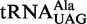 ,
, 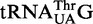 and
and 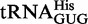 . (A) Selected yeast mitochondrial tRNAs with UAG or GUG anticodons (anticodon marked orange) are aligned (upper and lower blocks of sequences, respectively). Square brackets indicate the four standard helical regions in tRNAs. The arrow points to a nucleotide insertion that leads to a characteristic 8-nt anticodon loop in
. (A) Selected yeast mitochondrial tRNAs with UAG or GUG anticodons (anticodon marked orange) are aligned (upper and lower blocks of sequences, respectively). Square brackets indicate the four standard helical regions in tRNAs. The arrow points to a nucleotide insertion that leads to a characteristic 8-nt anticodon loop in 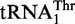 (
(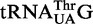 ). In A. gossypii, the anticodon loop has 7 nucleotides, and this tRNA reads CUN codons as Ala but not Thr. As
). In A. gossypii, the anticodon loop has 7 nucleotides, and this tRNA reads CUN codons as Ala but not Thr. As 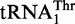 is most likely derived from tRNAHis by duplication, tRNAHis is included in the alignment for comparison [see also (21)]. Note the characteristic G residue at position −1 (constituting the 5′-terminus of tRNAHis) that pairs with the C at the 3′-discriminator position (both positions marked in gray). (B) Secondary structures of tRNAs that illustrate a possible tRNA reassignment scenario, S. cerevisiae
is most likely derived from tRNAHis by duplication, tRNAHis is included in the alignment for comparison [see also (21)]. Note the characteristic G residue at position −1 (constituting the 5′-terminus of tRNAHis) that pairs with the C at the 3′-discriminator position (both positions marked in gray). (B) Secondary structures of tRNAs that illustrate a possible tRNA reassignment scenario, S. cerevisiae
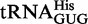 (left), A. gossypii
(left), A. gossypii
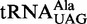 (right) and S. cerevisiae
(right) and S. cerevisiae
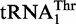 (middle). Nucleotide identity is coded as follows: red, A. gossypii
(middle). Nucleotide identity is coded as follows: red, A. gossypii
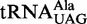 versus S. cerevisiae
versus S. cerevisiae
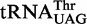 ; magenta, identity across all three tRNAs; and blue, additional identity between A. gossypii and S. cerevisiae
; magenta, identity across all three tRNAs; and blue, additional identity between A. gossypii and S. cerevisiae
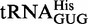 . Following a codon capture mechanism,
. Following a codon capture mechanism, 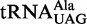 and
and 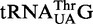 would have evolved from a
would have evolved from a 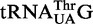 because the latter is present in the sister lineage Lachancea species (for a species phylogeny, see Figure 5), and absent in K. lactis. For abbreviations, see Figure 1.
because the latter is present in the sister lineage Lachancea species (for a species phylogeny, see Figure 5), and absent in K. lactis. For abbreviations, see Figure 1.