Figure 2.
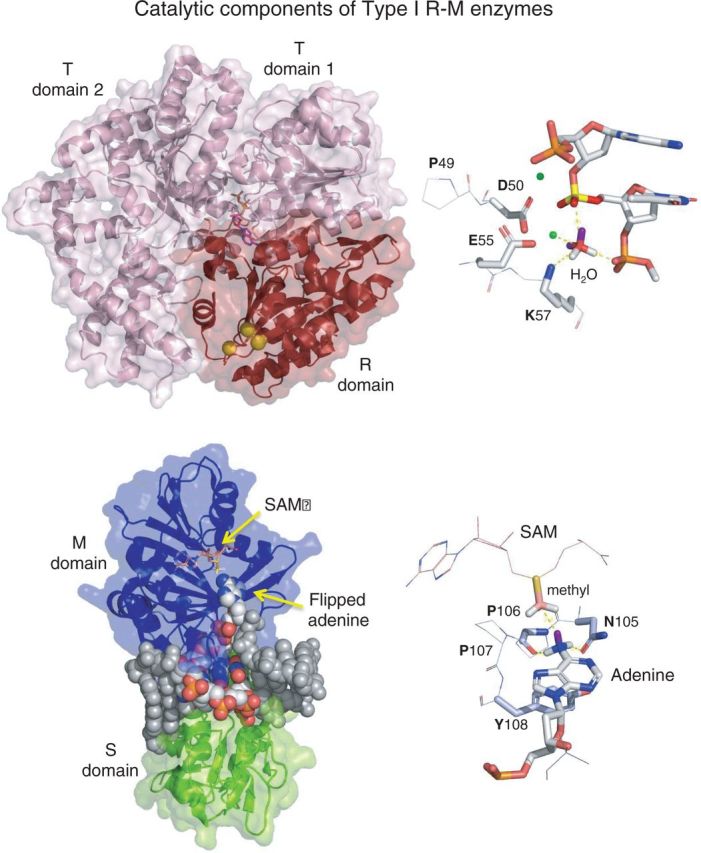
Catalytic components of Type I R–M enzymes. Upper figure, left: The R/T subunit of EcoR124I (pdb: 2W00). The R domain (red), responsible for DNA cleavage, comprises the N-terminal ∼260 aa. It contains the common PD-D/EXK endonuclease catalytic site, composed of D151, E165 and K167 (yellow spheres). Upper figure, right. The PD-D/EXK catalytic site of the Type II REase, MvaI (pdb:2OAA). D50 and E55 coordinate divalent metal ions (in this case two Ca2+ ions, shown as green spheres at reduced scale for clarity). The hydrolytic water molecule is oriented by interaction with a metal ion, the general base K57, and a phosphate oxygen from the adjacent base. These interactions position a lone pair electron orbital (purple sticks) of the water molecule for in-line nucleophilic attack on the phosphorus atom (bright yellow), initiating the DNA cleavage reaction. Catalysis occurs in the presence of Mg2+, but not in the presence of Ca2+; hence, this structure represents the pre-cleavage complex. Lower figure, left: The monomeric γ-MTase, M.TaqI, with bound DNA (pdb: 1G38). SAM was absent in this complex, which represents the pre-methylation complex. SAM has been added here through modeling by structural alignment with pdb:2ADM. Lower figure, right: The catalytic NPPY of M.TaqI is composed of N105, P106, P107 and Y108. When the target adenine is flipped into the catalytic site, the hydrogens of the 6-amino group form hydrogen bonds with the side chain amide carbonyl of N104 and the main chain carbonyl of P105. These lie below the plane of the base and likely induce the nitrogen to switch from the planar sp2 orbital configuration it normally possesses, to the tetrahedral sp3 configuration (105). In this latter configuration, the lone pair orbital of nitrogen (purple stick) is appropriately positioned for in-line nucleophilic attack on the carbon thiol (pink) of SAM, initiating the DNA methylation reaction (105,106).
