Figure 2.
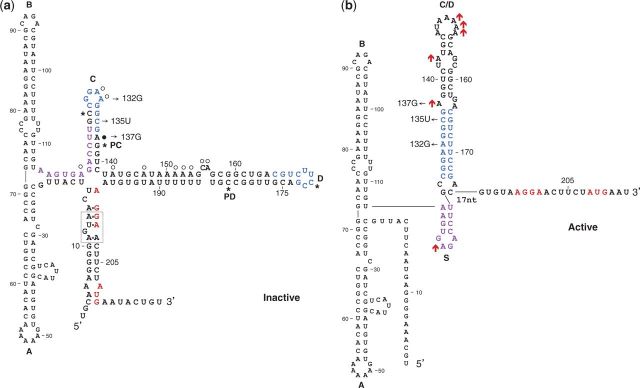
Translationally inactive (a) and active (b) structures. The Shine–Dalgarno and the initiation codon of alx are in red. Complementary sequences in hairpins C and D are in blue. Thin arrows indicate specific nucleotide changes generated by site-directed mutagenesis. To form the translationally active structure, the proximal part of hairpin C folds into the small hairpin S (nucleotides marked in purple), whereas basepairing between loops C and D forms stem C/D. A loop E motif in the inactive structure is boxed (dashed line). PC and PD denote the pause sites at hairpins C and D, respectively. Asterisks in (a) delineate the regions typically sequestered within the paused transcription elongation complex. The cotranscriptional structure probing data obtained on synthesis with E. coli RNAP (Figure 5) is presented on the structures. (a) Data obtained with wild-type RNA. Circles indicate strong (filled circles) and weak (open circles) DMS modification sites. (b) Data obtained with the high-level expression mutant A132G. Red upward arrows indicate increased modification intensity compared with wild-type. The two structures, the inactive and the active one, were previously shown to form under neutral and high pH conditions, respectively.
