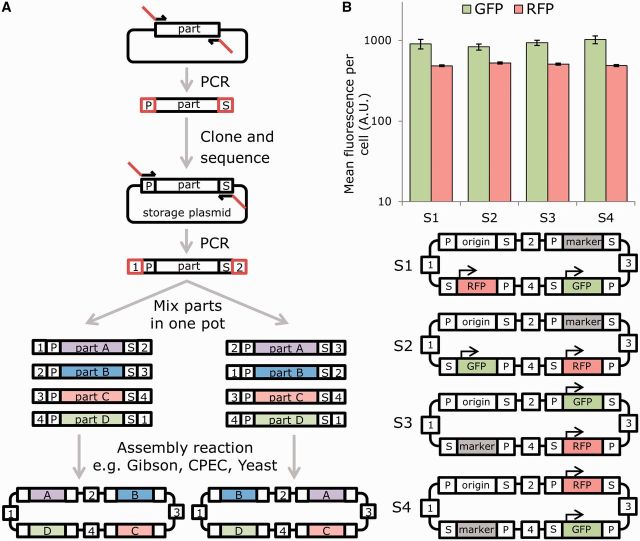
Figure 1.
Overlap-directed assembly of DNA parts by addition of modular linker sequences can enable standardized assembly of genes in different orders and orientations without context effects. (A) Schematic of the MODAL strategy. Using an initial PCR, selected DNA parts are amplified from their source to be flanked by defined 15-bp adapter sequences (P = prefix adapter, S = suffix adapter), which can then be cloned, stored and sequence verified in the pJET vector. Linker sequences (numbered 1–4) are then added 5′ and 3′ of the adapters by PCR and these guide homology-mediated overlap assembly into plasmids or other constructs. The bifurcation in the diagram shows an example of how changing which linkers are added to part A and part B changes their order in the final construct. (B) Using a single set of four random 45-bp linkers and four standardized E. coli parts (constitutive GFP expression, constitutive RFP expression, kanamycin resistance and a pUC origin of replication), Gibson assembly was used to construct plasmids with parts in a variety of orders. GFP and RFP expression per cell as measured by flow cytometry did not show significant variation when parts were shuffled to different positions in the plasmid and contained different linker sequences upstream and downstream of genes. Mean fluorescence per cell was calculated from mean FL1 (GFP) and mean FL5 (RFP) measurements (n = 5). Error bars indicate standard error.