Figure 2.
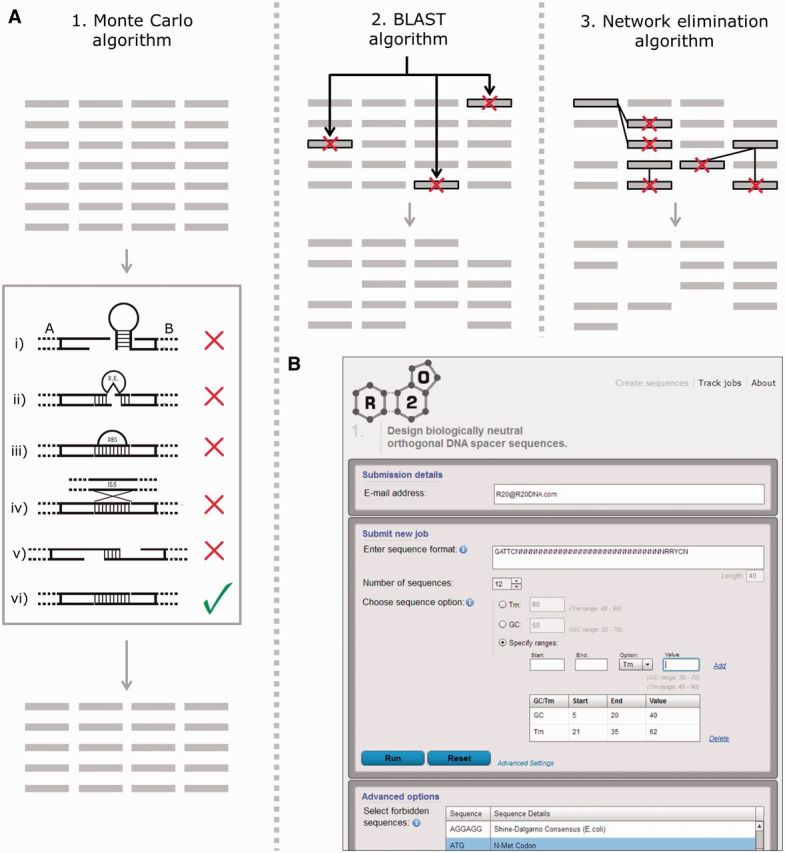
R2oDNA Designer is an online software tool for the design of synthetic linker sequences for the MODAL strategy. (A) Process overview. An initial pool of random sequences are generated that match primary user requirements (length- and position-specific nucleotide constraints). These are then optimized using MCSA using a scoring function that penalizes (i) single-stranded DNA secondary structures, (ii) forbidden sequence motifs including restriction enzyme sites, (iii) functional motifs like ribosome binding sequences and (iv) insertion element sequences, (v) unwanted self-annealing and (vi) off-target GC content or Tm. All sequences containing forbidden motifs or medium to strong secondary structures or self-annealing are optimized out. Any sequences with significant BLAST hits to selected genomes are eliminated. Finally, any remaining sequences that cross-anneal to one another are removed using a network elimination algorithm. (B) R2oDNA Designer user interface is shown. Users can input linker length, sequence requirements and preferences for GC content or melting temperature in the first window. A list of the forbidden sequences and genomes to compare with can be adjusted in the advanced settings window.
