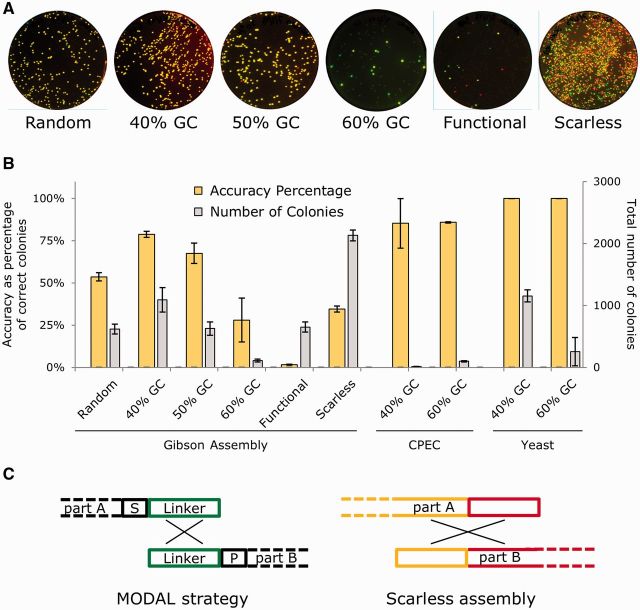
Figure 3.
Assessment of designed linkers used within the MODAL strategy with Gibson, CPEC and S. cerevisiae recombination DNA assembly methods. (A) To assess modular linker sequences, four parts were assembled by Gibson assembly into a plasmid encoding constitutive GFP and RFP expression, following the scheme illustrated in Figure 1A. DH10B E. coli were transformed with assembly reactions and grown on LB + kanamycin agar plates overnight and then scanned for green and red fluorescence the following day. This was repeated three times on separate days, and a single set is shown here. Assembly was done using six different linker sets: random, designed with 40% GC content, designed with 50% GC content, designed with 40% GC content, functional and scarless. Correct assembly gives colonies appearing yellow due to simultaneous green and red fluorescence. (B) The total number of colonies and the percentage of those containing correctly assembled plasmids (the ‘accuracy’) were calculated from image analysis of each plate for DNA assemblies using different linker sequences and using the Gibson, CPEC and S. cerevisiae recombination DNA assembly methods. Equivalent E. coli parts and competent cells were used for Gibson and CPEC (n = 3), but for S. cerevisiae assembly, YPH500 cells were used and DNA parts encoded constitutive yEGFP and mCherry RFP expression, uracil selection and a 2 -μ plasmid origin (n = 2). Error bars indicate standard error. (C) Schematic illustrating the difference between the assembly process and intergenic regions formed by the MODAL strategy and scarless assembly.