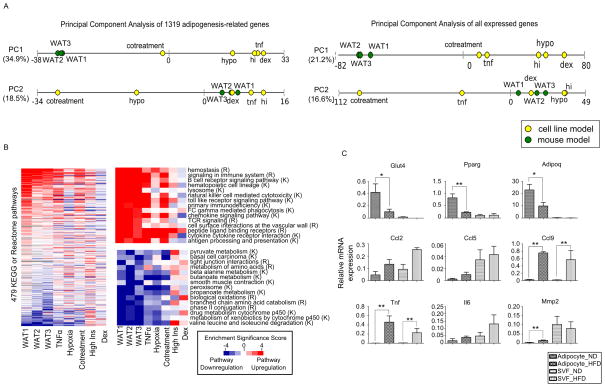
Figure 3. Systemic transcriptome changes in adipose insulin resistance revealed by global pathway analysis, See also Figure S4 and Tables S2, S3 and S4.
(A) Principal components analysis (PCA) of the eight models. Each principal component (PC1 and PC2) represents a direction of maximal variation in the matrix of expression data. The models are projected onto the first two principal components, with the cell line models shown as yellow dots and the mouse models shown as green dots. The distance between dots on each line corresponds to the distance between models along this projection. The numbers in parentheses represent the fraction of the variance in the expression fold change matrix that is explained by that particular principal component. Left: PCA using the 1,319 adipogenesis-related genes. Right: PCA using 13,043 genes with FPKM > 0.1.
(B) Heatmap showing the results from gene set enrichment analysis (GSEA) of genome-wide expression data. We computed the p-value for the significance of enrichment for either upregulation or downregulation of each pathway in the KEGG and Reactome databases. The log transformed p-values were taken as the Enrichment Significance Scores: upregulated pathways are represented as red (positive Enrichment Significance Score) and downregulated pathways as blue (negative Enrichment Significance Score), with the intensity of the color representing the significance of upregulation/downregulation; white represents no upregulation/downregulation; grey indicates that the number of genes was less than 15. Left: Rows (pathways) were ranked according to the sum of the Enrichment Significance Scores of the 3 mouse models. The scale bar is indicated at the right bottom corner of the figure and is the same for all panels. Top right: Illustration of the top 15 pathways upregulated in vivo (ranked as in the left panel). Pathways from Reactome are labeled (R) and those from KEGG are labeled (K). Bottom right: Illustration of the top 15 pathways downregulated in vivo.
(C) Expression of selected adipocyte and inflammatory response-related genes in isolated adipocytes or stromal vascular fraction (SVF) from normal chow-fed mice (ND) or high fat diet-fed mice (HFD) measured by qPCR. Relative expression is calculated by normalizing with the house keeping gene ribosomal protein S27 (Rps27). Data is presented as mean ± SEM (n=3). Statistical significance is indicated (*p < 0.05, **p < 0.01).