Figure 3. Abundance estimation via Expectation Maximization by RSEM.
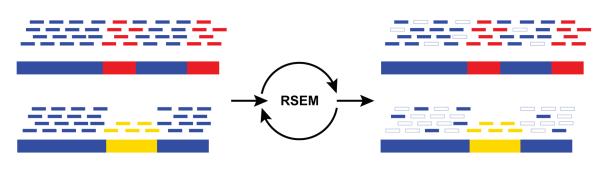
Shown is an illustrative example of abundance estimation for two transcripts with shared (blue) and unique (red, yellow) sequences. To estimate transcript abundances, RNA-Seq reads (short bars) are first aligned to the transcript sequences (long bars, bottom). Unique regions of isoforms will capture uniquely-mapping RNA-Seq reads (red and yellow short bars), and shared sequences between isoforms will capture multiply-mapping reads (blue short bars). An Expectation Maximization algorithm, implemented in the RSEM software, estimates the most likely relative abundances of the transcripts and then fractionally assigns reads to the isoforms based on these abundances. The assignments of reads to isoforms resulting from iterations of expectation maximization are illustrated as filled short bars (right), and those assignments eliminated are shown as hollow. Note that assignments of multiply-mapped reads are in fact performed fractionally according to a maximum likelihood estimate. Thus, in this example, a higher fraction of each read is assigned to the more highly expressed top isoform than to the bottom isoform.
