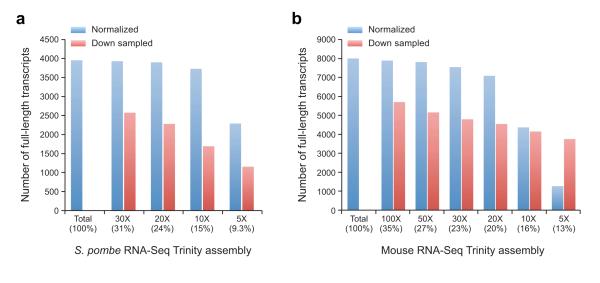
Figure 4. Effects of in silico fragment normalization of RNA-Seq data on Trinity full-length transcript reconstruction.
Shown are the number of full-length transcripts reconstructed (Y axis) from a dataset of paired-end strand-specific RNA-Seq in S. pombe (a, 10M paired-end reads) or mouse (b, 100M, paired-end reads), using either the full dataset (Total; 100%) or different samplings (X axis) by either Trinity’s in silico normalization procedure at 5X up to 100X targeted maximum k-mer (k=25) coverage (blue bars) or by random down-sampling of the same number of reads (red bars).