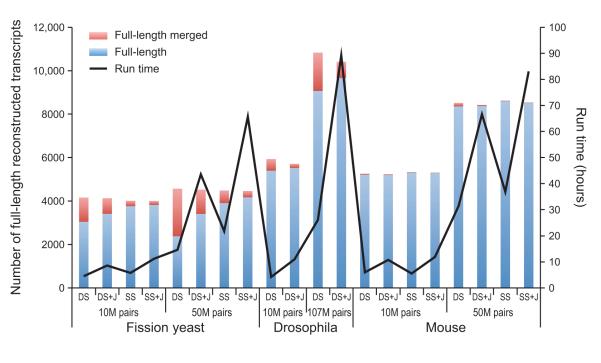
Figure 7. Full-length transcript reconstruction by Trinity in different organisms, sequencing depths, and parameters.
Shown is the number of fully reconstructed transcripts (bars, left Y axis) for Trinity assemblies of RNA-Seq data derived from fission yeast (Schizosaccharomyces pombe8, 45), Drosophila melanogaster11, and mouse8 with different combinations of parameters: DS – double stranded mode, SS – strand-specific mode, +J – using the ‘--jaccard_clip’ parameter to split falsely fused transcripts. Both SS and DS results are provided for S. pombe and mouse, but only DS results are provided for Drosophila since its RNA-Seq data was not strand-specific. Blue: full-length transcripts; red: full length merged, i.e., transcripts erroneously fused with another (typically neighboring) transcript. The black curve (right Y axis) indicates the run times in each case with a contemporary high-memory (256G to 512G RAM) server using a maximum of 4 threads (‘--CPU 4’, see Tutorial).